Abstract
The vesicular transport pathways, which shuttle materials to and from the cell surface and within the cell, and the metabolic (growth factor and nutrient) signalling pathways, which integrate a variety of extracellular and intracellular signals to mediate growth, proliferation or survival, are both important for cellular physiology. There is evidence to suggest that the transport and metabolic signalling pathways intersect—vesicular transport can affect the regulation of metabolic signals and vice versa. The Rab family GTPases regulate the specificity of vesicular transport steps in the cell. Together with their interacting proteins, Rabs would likely constitute the points of intersection between vesicular transport and metabolic signalling pathways. Examples of these points would include growth factor signalling, glucose and lipid metabolism, as well as autophagy. Many of these processes involve mechanistic/mammalian target of rapamycin (mTOR) complex 1 (mTORC1) in downstream cascades, or are regulated by TORC signalling. A general functionality of the vesicular transport processes controlled by the Rabs is also important for spatial and temporal regulation of the transmission of metabolic signals between the cell surface and the nucleus. In other cases, specific Rabs and their interacting proteins are known to function in recruiting metabolism-related proteins to target membranes, or may compete with other factors in the TORC signalling pathway as a means of metabolic regulation. We review and discuss herein examples of how Rabs and their interacting proteins can mediate metabolic signalling and regulation in cells.


Similar content being viewed by others
References
Segev N (2001) Ypt/rab gtpases: regulators of protein trafficking. Sci STKE 2001:RE11
Hutagalung AH, Novick PJ (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91:119–149
Ullrich O et al (1993) Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem 268:18143–18150
Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR (1997) Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J 16:465–472
Ohya T et al (2009) Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature 459:1091–1097
Grosshans BL, Ortiz D, Novick P (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA 103:11821–11827
Markgraf DF, Peplowska K, Ungermann C (2007) Rab cascades and tethering factors in the endomembrane system. FEBS Lett 581:2125–2130
Seabra MC, Coudrier E (2004) Rab GTPases and myosin motors in organelle motility. Traffic 5:393–399
Nottingham RM, Pfeffer SR (2009) Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci USA 106:14185–14186
Sorkin A, von Zastrow M (2009) Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 10:609–622
Howe CL (2005) Modeling the signaling endosome hypothesis: why a drive to the nucleus is better than a (random) walk. Theor Biol Med Model 2:43
Dinneen JL, Ceresa BP (2004) Expression of dominant negative rab5 in HeLa cells regulates endocytic trafficking distal from the plasma membrane. Exp Cell Res 294:509–522
Chen PI, Kong C, Su X, Stahl PD (2009) Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J Biol Chem 284:30328–30338
Ceresa BP (2006) Regulation of EGFR endocytic trafficking by rab proteins. Histol Histopathol 21:987–993
Kauppi M, Simonsen A, Bremnes B (2002) The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci 115:899–911
Ng EL, Ng JJ, Liang F, Tang BL (2009) Rab22B is expressed in the CNS astroglia lineage and plays a role in epidermal growth factor receptor trafficking in A431 cells. J Cell Physiol 221:716–728
Chua CEL, Tang BL (2014) Engagement of the small GTPase Rab31 protein and its effector, early endosome antigen 1, is important for trafficking of the ligand-bound epidermal growth factor receptor from the early to the late endosome. J Biol Chem 289:12375–12389
Ceresa BP, Bahr SJ (2006) Rab7 activity affects epidermal growth factor: epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem 281:1099–1106
Cullis DN, Philip B, Baleja JD, Feig LA (2002) Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem 277:49158–49166
De Renzis S, Sönnichsen B, Zerial M (2002) Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. (3). Nat Cell Biol 4:124–133
Chia WJ, Tang BL (2009) Emerging roles for Rab family GTPases in human cancer. Biochim Biophys Acta 1795:110–116
Tomshine JC et al (2009) Cell proliferation and epidermal growth factor signaling in non-small cell lung adenocarcinoma cell lines are dependent on Rin1. (1). J Biol Chem 284:26331–26339
Kotzsch M et al (2008) Urokinase receptor splice variant uPAR-del4/5-associated gene expression in breast cancer: identification of rab31 as an independent prognostic factor. Breast Cancer Res Treat 111:229–240
Wang Y, Pennock S, Chen X, Wang Z (2002) Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol Cell Biol 22:7279–7290
Wang L, Liang Z, Li G (2011) Rab22 controls NGF signaling and neurite outgrowth in PC12 cells. Mol Biol Cell 22:3853–3860
Ueno H, Huang X, Tanaka Y, Hirokawa N (2011) KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev Cell 20:60–71
Mao X et al (2006) APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol 8:516–523
Schenck A et al (2008) The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell 133:486–497
Tan Y, You H, Wu C, Altomare DA, Testa JR (2010) Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor-selective effects on Akt signaling in murine embryonic fibroblasts. J Biol Chem 285:6377–6389
Banach-Orlowska M, Pilecka I, Torun A, Pyrzynska B, Miaczynska M (2009) Functional characterization of the interactions between endosomal adaptor protein APPL1 and the NuRD co-repressor complex. Biochem J 423:389–400
Miaczynska M et al (2004) APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. (1). Cell 116:445–456
Scheepers A, Joost H, Schurmann A (2004) The glucose transporter families SGLT and GLUT: molecular basis of normal and aberrant function. J Parenter Enter Nutr 28:364–371
Wood IS, Trayhurn P (2003) Glucose transporters (GLUT and SGLT): expanded families of sugar transport proteins. Br J Nutr 89:3–9
Olson AL, Pessin JE (1996) Structure, function, and regulation of the mammalian facilitative glucose transporter gene family. Annu Rev Nutr 16:235–256
Thorens B, Sarkar HK, Kaback HR, Lodish HF (1988) Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and beta-pancreatic islet cells. Cell 55:281–290
Nagamatsu S et al (1992) Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J Biol Chem 267:467–472
Birnbaum MJ (1989) Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell 57:305–315
Zeigerer A et al (2002) GLUT4 retention in adipocytes requires two intracellular insulin-regulated transport steps. Mol Biol Cell 13:2421–2435
Rea S, James DE (1997) Moving GLUT4: the biogenesis and trafficking of GLUT4 storage vesicles. Diabetes 46:1667–1677
Sano H et al (2007) Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab 5:293–303
Mîinea CP et al (2005) AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J 391:87–93
Larance M (2005) Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J Biol Chem 280:37803–37813
Huang J, Imamura T, Olefsky JM (2001) Insulin can regulate GLUT4 internalization by signaling to Rab5 and the motor protein dynein. Proc Natl Acad Sci 98:13084–13089
Lodhi IJ et al (2007) Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab 5:59–72
Imamura T et al (2003) Insulin-induced GLUT4 translocation involves protein kinase C-lambda-mediated functional coupling between Rab4 and the motor protein kinesin. Mol Cell Biol 23:4892–4900
Li L (2001) Direct interaction of Rab4 with syntaxin 4. J Biol Chem 276:5265–5273
Kane S et al (2002) A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem 277:22115–22118
Sano H (2003) Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem 278:14599–14602
Leney SE, Tavaré JM (2009) The molecular basis of insulin-stimulated glucose uptake: signalling, trafficking and potential drug targets. J Endocrinol 203:1–18
Ishikura S, Bilan PJ, Klip A (2007) Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem Biophys Res Commun 353:1074–1079
Sun Y, Bilan PJ, Liu Z, Klip A (2010) Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci 107:19909–19914
Ishikura S, Klip A (2008) Muscle cells engage Rab8A and myosin Vb in insulin-dependent GLUT4 translocation. Am J Physiol Cell Physiol 295:C1016–C1025
Sun Y, Chiu TT, Foley KP, Bilan PJ, Klip A (2014) Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells. Mol Biol Cell 25:1159–1170
Ishikura S, Koshkina A, Klip A (2008) Small G proteins in insulin action: Rab and Rho families at the crossroads of signal transduction and GLUT4 vesicle traffic. Acta Physiol (Oxf) 192:61–74
Sadacca LA, Bruno J, Wen J, Xiong W, McGraw TE (2013) Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs. Mol Biol Cell 24:2544–2557
Reed SE et al (2013) A role for Rab14 in the endocytic trafficking of GLUT4 in 3T3-L1 adipocytes. J Cell Sci 126:1931–1941
Chen Y et al (2012) Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J Cell Biol 198:545–560
Chavez JA, Roach WG, Keller SR, Lane WS, Lienhard GE (2008) Inhibition of GLUT4 translocation by Tbc1d1, a Rab GTPase-activating protein abundant in skeletal muscle, is partially relieved by AMP-activated protein kinase activation. J Biol Chem 283:9187–9195
Szekeres F et al (2012) The Rab-GTPase-activating protein TBC1D1 regulates skeletal muscle glucose metabolism. Am J Physiol Endocrinol Metab 303:E524–E533
Peck GR et al (2009) Insulin-stimulated phosphorylation of the Rab GTPase-activating protein TBC1D1 regulates GLUT4 translocation. J Biol Chem 284:30016–30023
Dokas J et al (2013) Conventional knockout of Tbc1d1 in mice impairs insulin- and AICAR-stimulated glucose uptake in skeletal muscle. Endocrinology 154:3502–3514
Lansey MN, Walker NN, Hargett SR, Stevens JR, Keller SR (2012) Deletion of Rab GAP AS160 modifies glucose uptake and GLUT4 translocation in primary skeletal muscles and adipocytes and impairs glucose homeostasis. Am J Physiol Endocrinol Metab 303:E1273–E1286
Chadt A et al (2014) Deletion of both Rab-GTPase-activating proteins TBC1D1 and TBC1D4 in mice eliminates insulin- and AICAR-stimulated glucose transport. Diabetes. doi:10.2337/db14-0368
Yamauchi T et al (2002) Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 8:1288–1295
Yamauchi T et al (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423:762–769
Ding Q, Wang Z, Chen Y (2009) Endocytosis of adiponectin receptor 1 through a clathrin- and Rab5-dependent pathway. Cell Res 19:317–327
Deepa SS, Dong LQ (2008) APPL1: role in adiponectin signaling and beyond. AJP Endocrinol Metab 296:E22–E36
Wang C et al (2009) Yin-Yang regulation of adiponectin signaling by APPL isoforms in muscle cells. (1). J Biol Chem 284:31608–31615
Cheng KKY et al (2014) The adaptor protein APPL2 inhibits insulin-stimulated glucose uptake by interacting with TBC1D1 in skeletal muscle. Diabetes 63:3748–3758
King GJ et al (2012) Membrane curvature protein exhibits interdomain flexibility and binds a small GTPase. J Biol Chem 287:40996–41006
Thiam AR, Farese RV, Walther TC (2013) The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14:775–786
Krahmer N, Farese RV, Walther TC (2013) Balancing the fat: lipid droplets and human disease. EMBO Mol Med 5:905–915
Stehr M, Elamin AA, Singh M (2012) Cytosolic lipid inclusions formed during infection by viral and bacterial pathogens. Microbes Infect 14:1227–1237
Liu P et al (2004) Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem 279:3787–3792
Brasaemle DL, Dolios G, Shapiro L, Wang R (2004) Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem 279:46835–46842
Turró S et al (2006) Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic 7:1254–1269
Kiss RS, Nilsson T (2014) Rab proteins implicated in lipid storage and mobilization. J Biomed Res 28:169–177
Wilfling F, Haas JT, Walther TC, Farese RV (2014) Lipid droplet biogenesis. Curr Opin Cell Biol 29:39–45
Robenek MJ et al (2004) Lipids partition caveolin-1 from ER membranes into lipid droplets: updating the model of lipid droplet biogenesis. FASEB J Off Publ Fed Am Soc Exp Biology 18:866–868
Robenek H et al (2006) Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci 119:4215–4224
Nevo-Yassaf I et al (2012) Role for TBC1D20 and Rab1 in hepatitis C virus replication via interaction with lipid droplet-bound nonstructural protein 5A. J Virol 86:6491–6502
Liu P et al (2007) Rab-regulated interaction of early endosomes with lipid droplets. Biochim Biophys Acta 1773:784–793
Weidberg H, Shvets E, Elazar Z (2009) Lipophagy: selective catabolism designed for lipids. Dev Cell 16:628–630
Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG (2005) Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem 280:42325–42335
Pulido MR et al (2011) Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One 6:e22931
Boström P et al (2007) SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol 9:1286–1293
Salloum S, Wang H, Ferguson C, Parton RG, Tai AW (2013) Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog 9:e1003513
Dansako H, Hiramoto H, Ikeda M, Wakita T, Kato N (2014) Rab18 is required for viral assembly of hepatitis C virus through trafficking of the core protein to lipid droplets. Virology 462–463:166–174
Vogt DA et al (2013) Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog 9:e1003302
Aivazian D, Serrano RL, Pfeffer S (2006) TIP47 is a key effector for Rab9 localization. J Cell Biol 173:917–926
Ploen D et al (2013) TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur J Cell Biol 92:374–382
You X et al (2013) Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis 34:1644–1652
Wu L et al (2014) Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Dev Cell 30:378–393
Xu L, Zhou L, Li P (2012) CIDE proteins and lipid metabolism. Arterioscler Thromb Vasc Biol 32:1094–1098
Gong J et al (2011) Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol 195:953–963
Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6:463–477
Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E (2005) Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84–91
Lipatova Z et al (2012) Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci 109:6981–6986
Fader CM, Sánchez D, Furlán M, Colombo MI (2008) Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9:230–250
Longatti A et al (2012) TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 197:659–675
Munafo DB, Colombo MI (2002) Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 3:472–482
Mizushima N, Noda T, Ohsumi Y (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 18:3888–3896
Itoh T et al (2008) Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 19:2916–2925
Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M (2011) OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 192:839–853
Popovic D et al (2012) Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 32:1733–1744
Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905–909
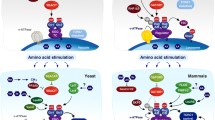
Zoncu R et al (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334:678–683
Jung CH, Ro SH, Cao J, Otto NM, Kim DH (2010) mTOR regulation of autophagy. FEBS Lett 584:1287–1295
Avruch J et al (2009) Activation of mTORC1 in two steps: rheb-GTP activation of catalytic function and increased binding of substrates to raptor1. Biochem Soc Trans 37:223–226
Sato T, Nakashima A, Guo L, Tamanoi F (2009) Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem 284:12783–12791
Dazert E, Hall MN (2011) mTOR signaling in disease. Curr Opin Cell Biol 23:744–755
Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell 149:274–293
Robida-Stubbs S et al (2012) TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 15:713–724
Kim DH et al (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–176
Sarbassov DD et al (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14:1296–1302
Inoki K, Li Y, Xu T, Guan KL (2003) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17:1829–1834
Cai SL (2006) Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol 173:279–289
Zoncu R et al (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. (1). Science 334:678–683
Sancak Y et al (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141:290–303
Kamada Y et al (2005) Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol 25:7239–7248
Cybulski N, Hall MN (2009) TOR complex 2: a signaling pathway of its own. Trends Biochem Sci 34:620–627
Sarbassov DD et al (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22:159–168
Li L et al (2010) Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem 285:19705–19709
Durán RV, Hall MN (2012) Regulation of TOR by small GTPases. EMBO Rep 13:121–128
Bridges D et al (2012) Rab5 proteins regulate activation and localization of target of rapamycin complex 1. (1). J Biol Chem 287:20913–20921
Roberts RL, Barbieri MA, Ullrich J, Stahl PD (2000) Dynamics of rab5 activation in endocytosis and phagocytosis. J Leukoc Biol 68:627–632
Matsui T, Fukuda M (2013) Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep 14:450–457
Luo L et al (2014) Rab8a interacts directly with PI3Kγ to modulate TLR4-driven PI3K and mTOR signalling. Nat Commun 5:4407
Tatebe H, Morigasaki S, Murayama S, Zeng CT, Shiozaki K (2010) Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr Biol 20:1975–1982
Tatebe H, Shiozaki K (2010) Rab small GTPase emerges as a regulator of TOR complex 2. Small GTPases 1:180–182
Martinez O, Schmidt A (1994) The small GTP-binding protein rab6 functions in intra-Golgi transport. J Cell Biol 127:1575–1588
He Y et al (2006) Genetic and functional interaction between Ryh1 and Ypt3: two Rab GTPases that function in S. pombe secretory pathway. Genes Cells 11:207–221
Marion RM (2004) Inaugural article: Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci 101:14315–14322
Fingerman I, Nagaraj V, Norris D, Vershon AK (2003) Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell 2:1061–1068
Lempiäinen H et al (2009) Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell 33:704–716
Singh J, Tyers M (2009) A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Genes Dev 23:1944–1958
Zhou QL et al (2008) Akt substrate TBC1D1 regulates GLUT1 expression through the mTOR pathway in 3T3-L1 adipocytes. Biochem J 411:647–655
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chua, C.E.L., Tang, B.L. Role of Rab GTPases and their interacting proteins in mediating metabolic signalling and regulation. Cell. Mol. Life Sci. 72, 2289–2304 (2015). https://doi.org/10.1007/s00018-015-1862-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-015-1862-x