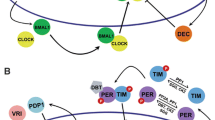
Abstract
An endogenous timing mechanism, the circadian clock, causes rhythmic expression of a considerable fraction of the genome of most organisms to optimally align physiology and behavior with their environment. Circadian clocks are self-sustained oscillators primarily based on transcriptional feedback loops and post-translational modification of clock proteins. It is increasingly becoming clear that regulation at the RNA level strongly impacts the cellular circadian transcriptome and proteome as well as the oscillator mechanism itself. This review focuses on posttranscriptional events, discussing RNA-binding proteins that, by influencing the timing of pre-mRNA splicing, polyadenylation and RNA decay, shape rhythmic expression profiles. Furthermore, recent findings on the contribution of microRNAs to orchestrating circadian rhythms are summarized.







Similar content being viewed by others
Abbreviations
- AANAT:
-
Serotonin N-acetyltransferase
- AGO1:
-
ARGONAUTE1
- ARE:
-
AU-rich element
- AS:
-
Alternative splicing
- bHLH:
-
Basic helix-loop-helix
- CCA1:
-
CIRCADIAN CLOCK ASSOCIATED1
- CIRP:
-
Cold-inducible RNA-binding protein
- CLK:
-
CLOCK
- CRY:
-
CRYPTHOCHROME
- CYC:
-
CYCLE
- FLC:
-
FLOWERING LOCUS C
- FMRP:
-
Fragile X mental retardation protein
- FRH:
-
FRQ-interacting helicase
- FRQ:
-
FREQUENCY
- GI:
-
GIGANTEA
- hnRNP:
-
Heterogenous nuclear ribonucleoprotein
- IRES:
-
Internal ribosome entry sites
- KH:
-
K homology
- LHY:
-
LATE ELONGATED HYPOCOTYL
- NAT:
-
Natural antisense transcripts
- NMD:
-
Nonsense-mediated decay
- PABP:
-
Poly(A) binding protein
- PDP:
-
PAR DOMAIN PROTEIN
- PER:
-
PERIOD
- PRR:
-
PSEUDO RESPONSE REGULATOR
- PTB:
-
Polypyrimidine tract-binding protein
- PTC:
-
Premature termination codon
- RBM:
-
RNA binding motif protein
- RBP:
-
RNA-binding protein
- RRM:
-
RNA recognition motif
- SCN:
-
Suprachiasmatic nuclei
- TF:
-
Transcription factor
- TIM:
-
TIMELESS
- TOC1:
-
TIMING OF CAB EXPRESSION 1
- UTR:
-
Untranslated region
- VRI:
-
VRILLE
- WC:
-
WHITE COLLAR
- WCC:
-
WHITE COLLAR Complex
References
Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U (2004) The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113:103–112
Mas P (2008) Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol 18:273–281
Schöning JC, Staiger D (2005) At the pulse of time: protein interactions determine the pace of circadian clocks. FEBS Lett 579:3246–3252
Gallego M, Virshup DM (2007) Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8:139–148
Mehra A, Baker CL, Loros JJ, Dunlap JC (2009) Post-translational modifications in circadian rhythms. Trends Biochem Sci 34:483–490
Edery I (1999) Role of post-transcriptional regulation in circadian clocks: lessons from Drosophila. Chronobiol Int 16:377–414
Harms E, Kivimae S, Young MW, Saez L (2004) Post-transcriptional and post-translational regulation of clock genes. J Biol Rhythms 19:361–373
Staiger D, Streitner C, Rudolf F, Huang X (2006) Multiple and slave oscillators, In: Hall A, McWatters H (eds) Endogenous plant rhythms. Blackwell, Oxford, pp 57–83
Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582:1977–1986
Moore MJ (2005) From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514–1548
Keene JD, Tenenbaum SA (2002) Eukaryotic mRNPs may represent post-transcriptional operons. Mol Cell 9:1161–1167
Tenenbaum SA, Carson CC, Lager PJ, Keene JD (2000) Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA 97:14085–14090
Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543
Keene JD (2007) Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol 72:157–165
Staiger D (2001) RNA-binding proteins and circadian rhythms in Arabidopsis thaliana. Philos Trans R Soc Lond B 356:1755–1759
Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417:78–83
Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi H, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by circadian clock. Cell 109:307–320
Pilgrim ML, Caspar T, Quail PH, McClung CR (1993) Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Mol Biol 23:349–364
Dibner C, Sage D, Unser M, Bauer C, d’Esmond T, Naef F, Schibler U (2009) Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28:123–134
Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH (2006) Circadian orchestration of the hepatic proteome. Curr Biol 16:1107–1115
Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96:271–290
Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6:544–556
Allada R, Chung BY (2010) Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72:605–624
Hardin PE (2005) The circadian timekeeping system of Drosophila. Curr Biol 15:R714–R722
Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J (2003) vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112:329–341
Dibner C, Schibler U, Albrecht U (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72:517–549
Ukai H, Ueda HR (2010) Systems biology of mammalian circadian clocks. Annu Rev Physiol 72:579–603
Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB (2006) Feedback repression is required for mammalian circadian clock function. Nat Genet 38:312–319
Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U (2002) The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251–260
Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K-I, Suzuki Y, Sugano S, Masamitsu I, Shigeyoshi Y, Hashimoto S (2002) A transcription factor response element for gene expression during circadian night. Nature 418:534–539
Brunner M, Kaldi K (2008) Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol 68:255–262
Locke JCW, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2:59
Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142:797–806
Schulze T, Prager K, Dathe H, Kelm J, Kiessling P, Mittag M (2010) How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma. doi:10.1007/s00709-010-0113-0
Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P (1992) The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci 16:123–127
Jacobshagen S, Kessler B, Rinehart CA (2008) At least four distinct circadian regulatory mechanisms are required for all phases of rhythms in mRNA amount. J Biol Rhythms 23:511–524
So WV, Rosbash M (1997) Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J 16:7146–7155
Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC (1994) A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12:555–570
Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC (1997) Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J 16:5006–5018
Mitchell P, Tollervey D (2000) mRNA stability in eukaryotes. Curr Opin Genet Dev 10:193–198
Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT (2009) Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res 37:26–37
Kwak E, Kim TD, Kim KT (2006) Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J Biol Chem 281:19100–19106
Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT (2010) Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol 30:197–205
Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT (2005) Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol Cell Biol 25:3232–3246
Kim TD, Woo K-C, Cho S, Ha D-C, Jang SK, Kim K-T (2007) Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev 21:797–810
Benjamin D, Schmidlin M, Min L, Gross B, Moroni C (2006) BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol Cell Biol 26:9497–9507
Cheng P, He Q, Wang L, Liu Y (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev 19:234–241
Guo J, Cheng P, Yuan H, Liu Y (2009) The exosome regulates circadian gene expression in a post-transcriptional negative feedback loop. Cell 138:1236–1246
Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99:11513–11518
Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138:2374–2385
Yakir E, Hilman D, Hassidim M, Green RM (2007) CCA1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol 145:925–932
Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127
Robinson BG, Frim DM, Schwartz WJ, Majzoub JA (1988) Vasopressin mRNA in the suprachiasmatic nuclei: daily regulation of polyadenylate tail length. Science 241:342–344
Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC (2001) Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol 1:9
Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC (2007) Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci USA 104:9888–9893
Cheng Y, Gvakharia B, Hardin PE (1998) Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol Cell Biol 18:6505–6514
Collins BH, Rosato E, Kyriacou CP (2004) Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci USA 101:1945–1950
Majercak J, Sidote D, Hardin PE, Edery I (1999) How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24:219–230
Majercak J, Chen WF, Edery I (2004) Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol 24:3359–3372
Low KH, Lim C, Ko HW, Edery I (2008) Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron 60:1054–1067
Liu Y, Garceau NY, Loros JJ, Dunlap JC (1997) Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell 89:477–486
Colot HV, Loros JJ, Dunlap JC (2005) Temperature-modulated alternative splicing and promoter use in the circadian clock gene frequency. Mol Biol Cell 16:5563–5571
Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D (1994) A light- and temperature-entrained circadian clock controls expression of transcripts encoding nuclear proteins with homology to RNA-binding proteins in meristematic tissue. Plant J 5:799–813
Carpenter CD, Kreps JA, Simon AE (1994) Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol 104:1015–1025
Staiger D, Apel K (1999) Circadian clock-regulated expression of an RNA-binding protein in Arabidopsis: characterisation of a minimal promoter element. Mol Gen Genet 261:811–819
Heintzen C, Nater M, Apel K, Staiger D (1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:8515–8520
Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33:361–371
Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D (2007) Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J 52:1119–1130
Schüttpelz M, Schöning JC, Doose S, Neuweiler H, Peters E, Staiger D, Sauer M (2008) Changes of conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J Am Chem Soc 130:9507–9513
Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36:6977–6987
McGlincy NJ, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33:385–393
Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR (2007) A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447:284–288
Kim JS, Jung HJ, Lee HJ, Kim KA, Goh C-H, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55:455–466
Schmidt F, Marnef A, Cheung M-K, Wilson I, Hancock J, Staiger D, Ladomery M (2010) A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol Biol Rep 37:839–845
Fujita J (1999) Cold shock response in mammalian cells. J Mol Microbiol Biotechnol 1:243–255
Nishiyama H, Xue JH, Sato T, Fukuyama H, Mizuno N, Houtani T, Sugimoto T, Fujita J (1998) Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem Biophys Res Commun 245:534–538
Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U (2007) System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5:e34
Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO, Kay SA (2009) Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol 10:R17
Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39:877–885
Newby LM, Jackson FR (1996) Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol 31:117–128
Huang Y, Genova G, Roberts M, Jackson FR (2007) The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE 2:e1107
Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, Botas J, Jackson FR, Nelson DL (2008) The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J Neurosci 28:10200–10205
Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, Albrecht U, Tamanini F, Meijer JH, Oostra BA, Nelson DL (2008) Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet 83:43–52
Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, Sakaki Y, Tei H (2007) LARK activates post-transcriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA 104:1859–1864
Höck J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G (2007) Proteomic and functional analysis of argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8:1052–1060
Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell 3:815–825
Waltenberger H, Schneid C, Grosch JO, Bareiss A, Mittag M (2001) Identification of target mRNAs for the clock-controlled RNA-binding protein Chlamy 1 from Chlamydomonas reinhardtii. Mol Genet Genomics 265:180–188
Kiaulehn S, Voytsekh O, Fuhrmann M, Mittag M (2007) The presence of UG-repeat sequences in the 3′-UTRs of reporter luciferase mRNAs mediates circadian expression and can determine acrophase in Chlamydomonas reinhardtii. J Biol Rhythms 22:275–277
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455:64–71
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114
Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW (2009) Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell 35:881–888
Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10:141–148
Yang M, Lee JE, Padgett RW, Edery I (2008) Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics 9:83
Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M (2009) A role for microRNAs in the Drosophila circadian clock. Genes Dev 23:2179–2191
Hipfner DR, Weigmann K, Cohen SM (2002) The bantam gene regulates Drosophila growth. Genetics 161:1527–1537
Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K (2007) microRNA modulation of circadian-clock period and entrainment. Neuron 54:813–829
Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D (2007) MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem 282:25053–25066
Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U (2009) Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 23:1313–1326
Sire C, Moreno AB, Garcia-Chapa M, Lopez-Moya JJ, Segundo BS (2009) Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett 583:1039–1044
Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic Flowering independent of CONSTANS in Arabidopsis. Plant Cell 19:2736–2748
Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK (2003) Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421:948–952
Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18:639–650
Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327:94–97
Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW 3rd, Janes J, Su AI, Hogenesch JB, Kay SA (2009) A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139:199–210
Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, Westhof E (2009) The RNA structure alignment ontology. RNA 15:1623–1631
Zong Q, Schummer M, Hood L, Morris DR (1999) Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci USA 96:10632–10636
Nandi A, Vaz C, Bhattacharya A, Ramaswamy R (2009) miRNA-regulated dynamics in circadian oscillator models. BMC Syst Biol 3:45
Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R, Croce CM, Calin GA, Perrotti D (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140:652–665
Acknowledgments
We thank Jay Dunlap for communication of results prior to publication, Stefan Janssen for help in preparing the figures and the anonymous reviewers for their suggestions on the manuscript. Tino Köster is a fellow of the German National Academic Foundation. Work in our laboratory is supported by the German Research Foundation through Grant STA 653/2 and the SFB 613.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Staiger, D., Köster, T. Spotlight on post-transcriptional control in the circadian system. Cell. Mol. Life Sci. 68, 71–83 (2011). https://doi.org/10.1007/s00018-010-0513-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0513-5