Abstract
Background
Atherosclerosis is a chronic inflammatory disease characterized by abnormal lipid deposition in the arteries. Programmed cell death is involved in the inflammatory response of atherosclerosis, but PANoptosis, as a new form of programmed cell death, is still unclear in atherosclerosis. This study explored the key PANoptosis-related genes involved in atherosclerosis and their potential mechanisms through bioinformatics analysis.
Methods
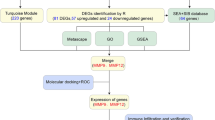
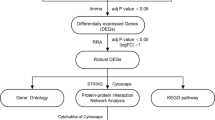
We evaluated differentially expressed genes (DEGs) and immune infiltration landscape in atherosclerosis using microarray datasets and bioinformatics analysis. By intersecting PANoptosis-related genes from the GeneCards database with DEGs, we obtained a set of PANoptosis-related genes in atherosclerosis (PANoDEGs). Functional enrichment analysis of PANoDEGs was performed and protein–protein interaction (PPI) network of PANoDEGs was established. The machine learning algorithms were used to identify the key PANoDEGs closely linked to atherosclerosis. Receiver operating characteristic (ROC) analysis was used to assess the diagnostic potency of key PANoDEGs. CIBERSORT was used to analyze the immune infiltration patterns in atherosclerosis, and the Spearman method was used to study the relationship between key PANoDEGs and immune infiltration abundance. The single gene enrichment analysis of key PANoDEGs was investigated by GSEA. The transcription factors and target miRNAs of key PANoDEGs were predicted by Cytoscape and online database, respectively. The expression of key PANoDEGs was validated through animal and cell experiments.
Results
PANoDEGs in atherosclerosis were significantly enriched in apoptotic process, pyroptosis, necroptosis, cytosolic DNA-sensing pathway, NOD-like receptor signaling pathway, lipid and atherosclerosis. Four key PANoDEGs (ZBP1, SNHG6, DNM1L, and AIM2) were found to be closely related to atherosclerosis. The ROC curve analysis demonstrated that the key PANoDEGs had a strong diagnostic potential in distinguishing atherosclerotic samples from control samples. Immune cell infiltration analysis revealed that the proportion of initial B cells, plasma cells, CD4 memory resting T cells, and M1 macrophages was significantly higher in atherosclerotic tissues compared to normal tissues. Spearman analysis showed that key PANoDEGs showed strong correlations with immune cells such as T cells, macrophages, plasma cells, and mast cells. The regulatory networks of the four key PANoDEGs were established. The expression of key PANoDEGs was verified in further cell and animal experiments.
Conclusions
This study evaluated the expression changes of PANoptosis-related genes in atherosclerosis, providing a reference direction for the study of PANoptosis in atherosclerosis and offering potential new avenues for further understanding the pathogenesis and treatment strategies of atherosclerosis.









Similar content being viewed by others
Data availability
The data in this study are publicly available in the Gene Expression Omnibus (GEO) database (GSE100927 and GSE40231). In addition, PANoptosis genes can be obtained in GeneCards (https://www.genecards.org).
Abbreviations
- DEGs:
-
Differentially expressed genes
- PANoDEGs:
-
Differentially expressed genes associated with PANoptosis
- ROC:
-
Receiver operating characteristic
- ASCVD:
-
Atherosclerotic cardiovascular disease
- AS:
-
Atherosclerosis
- PCD:
-
Programmed cell death
- NLRP3:
-
Nod-like receptor family pyrin domain containing 3
- GEO:
-
Gene expression omnibus
- LASSO:
-
Least absolute shrinkage and selector operation
- SVM-RFE:
-
Support vector machine-recursive feature elimination
- Limma:
-
Linear Model of Microarray Data
- PPI:
-
Protein–protein interaction
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- STRING:
-
Search Tool for the Retrieval of Interacting Genes
- AUC:
-
Area under curve
- GSEA:
-
Gene set enrichment analysis
- CTD:
-
Comparative toxicogenomics database
- TFs:
-
Transcription factors
- PMA:
-
Phorbol 12-myristate 13-acetate
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- CVD:
-
Cardiovascular disease
- ZBP1:
-
Z-DNA-Binding protein 1
- AIM2:
-
Melanoma 2
- DNM1L:
-
Dynamin-1-like protein
- DRP1:
-
Dynamin-related protein 1
References
Liu YT, Zhang ZM, Li ML, Gao S, Feng F, Xu WH. Association of carotid artery geometries with middle cerebral artery atherosclerosis. Atherosclerosis. 2022;352:27–34.
Sandesara PB, Virani SS, Fazio S, Shapiro MD. The forgotten lipids: triglycerides, remnant cholesterol, and atherosclerotic cardiovascular disease risk. Endocr Rev. 2019;40:537–57.
Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res. 2016;118:535–46.
Collaborators GDaI. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–22.
Colantonio LD, Shannon ED, Orroth KK, Zaha R, Jackson EA, Rosenson RS, Exter J, Mues KE, Muntner P. Ischemic event rates in very-high-risk adults. J Am Coll Cardiol. 2019;74:2496–507.
Li B, Xia Y, Hu B. Infection and atherosclerosis: TLR-dependent pathways. Cell Mol Life Sci CMLS. 2020;77:2751–69.
Zhang S, Li L, Chen W, Xu S, Feng X, Zhang L. Natural products: the role and mechanism in low-density lipoprotein oxidation and atherosclerosis. Phytothe Res PTR. 2021;35:2945–67.
Newton K, Dixit VM, Kayagaki N. Dying cells fan the flames of inflammation. Science (New York, NY). 2021;374:1076–80.
Malireddi RKS, Kesavardhana S, Kanneganti TD. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis). Front Cell Infect Microbiol. 2019;9:406.
Wang Y, Kanneganti TD. From pyroptosis, apoptosis and necroptosis to PANoptosis: a mechanistic compendium of programmed cell death pathways. Comput Struct Biotechnol J. 2021;19:4641–57.
Lee S, Karki R, Wang Y, Nguyen LN, Kalathur RC, Kanneganti TD. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature. 2021;597:415–9.
Paulin N, Viola JR, Maas SL, de Jong R, Fernandes-Alnemri T, Weber C, Drechsler M, Doring Y, Soehnlein O. Double-strand DNA sensing Aim2 inflammasome regulates atherosclerotic plaque vulnerability. Circulation. 2018;138:321–3.
Fidler TP, Xue C, Yalcinkaya M, Hardaway B, Abramowicz S, Xiao T, Liu W, Thomas DG, Hajebrahimi MA, Pircher J, et al. The AIM2 inflammasome exacerbates atherosclerosis in clonal haematopoiesis. Nature. 2021;592:296–301.
Weber C, Habenicht AJR, von Hundelshausen P. Novel mechanisms and therapeutic targets in atherosclerosis: inflammation and beyond. Eur Heart J. 2023;44:2672–81.
Mellby LD, Nyberg AP, Johansen JS, Wingren C, Nordestgaard BG, Bojesen SE, Mitchell BL, Sheppard BC, Sears RC, Borrebaeck CAK. Serum biomarker signature-based liquid biopsy for diagnosis of early-stage pancreatic cancer. J Clin Oncol. 2018;36:2887–94.
Fortino V, Wisgrill L, Werner P, Suomela S, Linder N, Jalonen E, Suomalainen A, Marwah V, Kero M, Pesonen M, et al. Machine-learning-driven biomarker discovery for the discrimination between allergic and irritant contact dermatitis. Proc Natl Acad Sci U S A. 2020;117:33474–85.
He Y, Zhang H, Ma L, Li J, Wang F, Zhou H, Zhang G, Wen Y. Identification of TIMP1 as an inflammatory biomarker associated with temporal lobe epilepsy based on integrated bioinformatics and experimental analyses. J Neuroinflammation. 2023;20:151.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43: e47.
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform. 2016;54:1–33.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–21.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. Correction to “The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets.” Nucleic Acids Res. 2021;49:10800.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22.
Lin X, Li C, Zhang Y, Su B, Fan M, Wei H. Selecting feature subsets based on SVM-RFE and the overlapping ratio with applications in bioinformatics. Molecules. 2017;23:52.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50.
Davis AP, Wiegers TC, Wiegers J, Wyatt B, Johnson RJ, Sciaky D, Barkalow F, Strong M, Planchart A, Mattingly CJ. CTD tetramers: a new online tool that computationally links curated chemicals, genes, phenotypes, and diseases to inform molecular mechanisms for environmental health. Toxicol Sci. 2023;195:155–68.
Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10:e1003731.
McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP. The biochemical basis of microRNA targeting efficacy. Science. 2019;366:1741.
Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–31.
Zheng Z, Yuan D, Shen C, Zhang Z, Ye J, Zhu L. Identification of potential diagnostic biomarkers of atherosclerosis based on bioinformatics strategy. BMC Med Genom. 2023;16:100.
Li M, Wang ZW, Fang LJ, Cheng SQ, Wang X, Liu NF. Programmed cell death in atherosclerosis and vascular calcification. Cell Death Dis. 2022;13:467.
Takahashi M. NLRP3 inflammasome as a key driver of vascular disease. Cardiovasc Res. 2022;118:372–85.
Fang X, Ardehali H, Min J, Wang F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat Rev Cardiol. 2023;20:7–23.
Puylaert P, Zurek M, Rayner KJ, De Meyer GRY, Martinet W. Regulated necrosis in atherosclerosis. Arterioscler Thromb Vasc Biol. 2022;42:1283–306.
Karki R, Kanneganti TD. ADAR1 and ZBP1 in innate immunity, cell death, and disease. Trends Immunol. 2023;44:201–16.
Zheng M, Kanneganti TD. The regulation of the ZBP1-NLRP3 inflammasome and its implications in pyroptosis, apoptosis, and necroptosis (PANoptosis). Immunol Rev. 2020;297:26–38.
Kuriakose T, Man SM, Malireddi RK, Karki R, Kesavardhana S, Place DE, Neale G, Vogel P, Kanneganti TD. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016;1:2045.
Kesavardhana S, Malireddi RKS, Burton AR, Porter SN, Vogel P, Pruett-Miller SM, Kanneganti TD. The Zalpha2 domain of ZBP1 is a molecular switch regulating influenza-induced PANoptosis and perinatal lethality during development. J Biol Chem. 2020;295:8325–30.
Kesavardhana S, Kuriakose T, Guy CS, Samir P, Malireddi RKS, Mishra A, Kanneganti TD. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J Exp Med. 2017;214:2217–29.
Place DE, Lee S, Kanneganti TD. PANoptosis in microbial infection. Curr Opin Microbiol. 2021;59:42–9.
Jiao H, Wachsmuth L, Kumari S, Schwarzer R, Lin J, Eren RO, Fisher A, Lane R, Young GR, Kassiotis G, et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature. 2020;580:391–5.
Karki R, Sundaram B, Sharma BR, Lee S, Malireddi RKS, Nguyen LN, Christgen S, Zheng M, Wang Y, Samir P, et al. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021;37:109858.
Karki R, Lee S, Mall R, Pandian N, Wang Y, Sharma BR, Malireddi RS, Yang D, Trifkovic S, Steele JA, et al. ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Sci Immunol. 2022;7:6294.
Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 2015;162:45–58.
Matyszewski M, Zheng W, Lueck J, Mazanek Z, Mohideen N, Lau AY, Egelman EH, Sohn J. Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome. Nat Commun. 2021;12:2735.
Du L, Wang X, Chen S, Guo X. The AIM2 inflammasome: A novel biomarker and target in cardiovascular disease. Pharmacol Res. 2022;186:106533.
Sharma BR, Karki R, Kanneganti TD. Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur J Immunol. 2019;49:1998–2011.
Kumari P, Russo AJ, Shivcharan S, Rathinam VA. AIM2 in health and disease: inflammasome and beyond. Immunol Rev. 2020;297:83–95.
Chou WC, Guo Z, Guo H, Chen L, Zhang G, Liang K, Xie L, Tan X, Gibson SA, Rampanelli E, et al. AIM2 in regulatory T cells restrains autoimmune diseases. Nature. 2021;591:300–5.
Hakimi M, Peters A, Becker A, Bockler D, Dihlmann S. Inflammation-related induction of absent in melanoma 2 (AIM2) in vascular cells and atherosclerotic lesions suggests a role in vascular pathogenesis. J Vasc Surg. 2014;59:794–803.
Tall AR, Bornfeldt KE. Inflammasomes and atherosclerosis: a mixed picture. Circ Res. 2023;132:1505–20.
Soehnlein O, Tall AR. AIMing 2 treat atherosclerosis. Nat Rev Cardiol. 2022;19:567–8.
Kraus F, Roy K, Pucadyil TJ, Ryan MT. Function and regulation of the divisome for mitochondrial fission. Nature. 2021;590:57–66.
Zeng Z, You M, Fan C, Rong R, Li H, Xia X. Pathologically high intraocular pressure induces mitochondrial dysfunction through Drp1 and leads to retinal ganglion cell PANoptosis in glaucoma. Redox Biol. 2023;62: 102687.
Quiles JM, Gustafsson AB. The role of mitochondrial fission in cardiovascular health and disease. Nat Rev Cardiol. 2022;19:723–36.
You Y, Chen X, Chen Y, Pang J, Chen Q, Liu Q, Xue H, Zeng Y, Xiao J, Mi J, et al. Epigenetic modulation of Drp1-mediated mitochondrial fission by inhibition of S-adenosylhomocysteine hydrolase promotes vascular senescence and atherosclerosis. Redox Biol. 2023;65:102828.
Wang L, Yu T, Lee H, O’Brien DK, Sesaki H, Yoon Y. Decreasing mitochondrial fission diminishes vascular smooth muscle cell migration and ameliorates intimal hyperplasia. Cardiovasc Res. 2015;106:272–83.
Umezu R, Koga JI, Matoba T, Katsuki S, Wang L, Hasuzawa N, Nomura M, Tsutsui H, Egashira K. Macrophage (Drp1) dynamin-related protein 1 accelerates intimal thickening after vascular injury. Arterioscler Thromb Vasc Biol. 2020;40:e214–26.
Rogers MA, Maldonado N, Hutcheson JD, Goettsch C, Goto S, Yamada I, Faits T, Sesaki H, Aikawa M, Aikawa E. Dynamin-related protein 1 inhibition attenuates cardiovascular calcification in the presence of oxidative stress. Circ Res. 2017;121:220–33.
Rogers MA, Hutcheson JD, Okui T, Goettsch C, Singh SA, Halu A, Schlotter F, Higashi H, Wang L, Whelan MC, et al. Dynamin-related protein 1 inhibition reduces hepatic PCSK9 secretion. Cardiovasc Res. 2021;117:2340–53.
Wang Y, Subramanian M, Yurdagul A Jr, Barbosa-Lorenzi VC, Cai B, de Juan-Sanz J, Ryan TA, Nomura M, Maxfield FR, Tabas I. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell. 2017;171(331–345):e322.
Cai G, Zhu Q, Yuan L, Lan Q. LncRNA SNHG6 acts as a prognostic factor to regulate cell proliferation in glioma through targeting p21. Biomed Pharmacother. 2018;102:452–7.
Wang HS, Zhang W, Zhu HL, Li QP, Miao L. Long noncoding RNA SNHG6 mainly functions as a competing endogenous RNA in human tumors. Cancer Cell Int. 2020;20:219.
Zhang X, Liu Z, Shu Q, Yuan S, Xing Z, Song J. LncRNA SNHG6 functions as a ceRNA to regulate neuronal cell apoptosis by modulating miR-181c-5p/BIM signalling in ischaemic stroke. J Cell Mol Med. 2019;23:6120–30.
Huang J, Jiang S, Liang L, He H, Liu Y, Cong L, Jiang Y. Analysis of PANoptosis-related LncRNA-miRNA-mRNA Network reveals LncRNA SNHG7 involved in chemo-resistance in colon adenocarcinoma. Front Oncol. 2022;12:888105.
Roy P, Orecchioni M, Ley K. How the immune system shapes atherosclerosis: roles of innate and adaptive immunity. Nat Rev Immunol. 2022;22:251–65.
Sun X, Yang Y, Meng X, Li J, Liu X, Liu H. PANoptosis: Mechanisms, biology, and role in disease. Immunol Rev. 2023;321:246–62.
Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019;25:1576–88.
Depuydt MAC, Prange KHM, Slenders L, Ord T, Elbersen D, Boltjes A, de Jager SCA, Asselbergs FW, de Borst GJ, Aavik E, et al. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res. 2020;127:1437–55.
Emoto T, Yamamoto H, Yamashita T, Takaya T, Sawada T, Takeda S, Taniguchi M, Sasaki N, Yoshida N, Saito Y, et al. Single-Cell RNA sequencing reveals a distinct immune landscape of myeloid cells in coronary culprit plaques causing acute coronary syndrome. Circulation. 2022;145:1434–6.
Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13:709–21.
Tabas I, Bornfeldt KE. Macrophage phenotype and function in different stages of atherosclerosis. Circ Res. 2016;118:653–67.
Luo Y, Duan H, Qian Y, Feng L, Wu Z, Wang F, Feng J, Yang D, Qin Z, Yan X. Macrophagic CD146 promotes foam cell formation and retention during atherosclerosis. Cell Res. 2017;27:352–72.
Tabas I, Lichtman AH. Monocyte-macrophages and T cells in atherosclerosis. Immunity. 2017;47:621–34.
Kuznetsova T, Prange KHM, Glass CK, de Winther MPJ. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat Rev Cardiol. 2020;17:216–28.
Xue S, Su Z, Liu D. Immunometabolism and immune response regulate macrophage function in atherosclerosis. Ageing Res Rev. 2023;90:101993.
Saigusa R, Winkels H, Ley K. T cell subsets and functions in atherosclerosis. Nat Rev Cardiol. 2020;17:387–401.
Freuchet A, Roy P, Armstrong SS, Oliaeimotlagh M, Kumar S, Orecchioni M, Ali AJ, Khan A, Makings J, Lyu Q, et al. Identification of human exT(reg) cells as CD16(+)CD56(+) cytotoxic CD4(+) T cells. Nat Immunol. 2023;24:1748–61.
Reilly NA, Lutgens E, Kuiper J, Heijmans BT, Jukema JW. Effects of fatty acids on T cell function: role in atherosclerosis. Nat Rev Cardiol. 2021;18:824–37.
Poznyak AV, Bezsonov EE, Popkova TV, Starodubova AV, Orekhov AN. Immunity in atherosclerosis: focusing on T and B cells. Int J Mol Sci. 2021;22:8379.
Hinkley H, Counts DA, VonCanon E, Lacy M. T Cells in atherosclerosis: key players in the pathogenesis of vascular disease. Cells. 2023;12:2152.
Kyaw T, Loveland P, Kanellakis P, Cao A, Kallies A, Huang AL, Peter K, Toh BH, Bobik A. Alarmin-activated B cells accelerate murine atherosclerosis after myocardial infarction via plasma cell-immunoglobulin-dependent mechanisms. Eur Heart J. 2021;42:938–47.
Taylor JA, Hutchinson MA, Gearhart PJ, Maul RW. Antibodies in action: the role of humoral immunity in the fight against atherosclerosis. Immun Ageing. 2022;19:59.
Otsuka F, Sakakura K, Virmani R. Are mast cells the real culprit in atherosclerosis? Eur Heart J. 2013;34:3681–3.
Wezel A, Lagraauw HM, van der Velden D, de Jager SC, Quax PH, Kuiper J, Bot I. Mast cells mediate neutrophil recruitment during atherosclerotic plaque progression. Atherosclerosis. 2015;241:289–96.
Kobayashi S, Kita S, Okuzaki D, Fujishima Y, Otsuki M, Kato H, Nishizawa Y, Miyashita K, Yokoyama C, Fukuhara A, et al. Favine/CCDC3 deficiency accelerated atherosclerosis and thrombus formation is associated with decreased MEF2C-KLF2 pathway. Science. 2022;25:105252.
Grootaert MOJ, Finigan A, Figg NL, Uryga AK, Bennett MR. SIRT6 protects smooth muscle cells from senescence and reduces atherosclerosis. Circ Res. 2021;128:474–91.
Chen Q, Lv J, Yang W, Xu B, Wang Z, Yu Z, Wu J, Yang Y, Han Y. Targeted inhibition of STAT3 as a potential treatment strategy for atherosclerosis. Theranostics. 2019;9:6424–42.
Liu H, Zuo C, Cao L, Yang N, Jiang T. Inhibition of miR-652-3p regulates lipid metabolism and inflammatory cytokine secretion of macrophages to alleviate atherosclerosis by improving TP53 expression. Mediators Inflamm. 2022;2022:9655097.
Nagao M, Lyu Q, Zhao Q, Wirka RC, Bagga J, Nguyen T, Cheng P, Kim JB, Pjanic M, Miano JM, Quertermous T. Coronary disease-associated gene TCF21 inhibits smooth muscle cell differentiation by blocking the myocardin-serum response factor pathway. Circ Res. 2020;126:517–29.
Lu YW, Martino N, Gerlach BD, Lamar JM, Vincent PA, Adam AP, Schwarz JJ. MEF2 (Myocyte Enhancer Factor 2) is essential for endothelial homeostasis and the atheroprotective gene expression program. Arterioscler Thromb Vasc Biol. 2021;41:1105–23.
Yung LM, Sanchez-Duffhues G, Ten Dijke P, Yu PB. Bone morphogenetic protein 6 and oxidized low-density lipoprotein synergistically recruit osteogenic differentiation in endothelial cells. Cardiovasc Res. 2015;108:278–87.
Cheng C, Tempel D, Den Dekker WK, Haasdijk R, Chrifi I, Bos FL, Wagtmans K, van de Kamp EH, Blonden L, Biessen EA, et al. Ets2 determines the inflammatory state of endothelial cells in advanced atherosclerotic lesions. Circ Res. 2011;109:382–95.
Xu F, Shen L, Chen H, Wang R, Zang T, Qian J, Ge J. circDENND1B participates in the antiatherosclerotic effect of IL-1beta monoclonal antibody in mouse by promoting cholesterol efflux via miR-17-5p/Abca1 Axis. Front Cell Dev Biol. 2021;9:652032.
Linna-Kuosmanen S, Tomas Bosch V, Moreau PR, Bouvy-Liivrand M, Niskanen H, Kansanen E, Kivela A, Hartikainen J, Hippelainen M, Kokki H, et al. NRF2 is a key regulator of endothelial microRNA expression under proatherogenic stimuli. Cardiovasc Res. 2021;117:1339–57.
Hong Q, Ling L, Huang W, Liu Y, Zhuo Y, Hong Z, Wu B, Zhang Y. LncRNA RNCR3 promotes endothelial cell proliferation and inflammatory cytokine secretion via regulating miR-185-5p/cyclin D2 axis. Environ Sci Pollut Res Int. 2021;28:27025–32.
Vyas HS, Jadeja RN, Vohra A, Upadhyay KK, Thounaojam MC, Bartoli M, Devkar RV. CORM-A1 alleviates pro-atherogenic manifestations via miR-34a-5p downregulation and an improved mitochondrial function. Antioxidants (Basel). 2023;12:997.
Zhang WB, Qi YF, Xiao ZX, Chen H, Liu SH, Li ZZ, Zeng ZF, Wu HF. CircHIPK3 regulates vascular smooth muscle cell calcification via the miR-106a-5p/MFN2 axis. J Cardiovasc Transl Res. 2022;15:1315–26.
Acknowledgements
We are grateful to all the contributors to the Gene Expression Omnibus (GEO) database, as well as the developers of the GeneCards database and the R packages we use.
Funding
This work was supported by 2022 Taizhou Science and Technology Support Program (Social Development) Project (No. TS202219), Clinical Specialist Talents' Professional Ability Innovation and Application Research Project (No. RCLX2315029) and 2022 Jiangsu Taizhou People's Hospital hospital-level Project (No. ZL202209).
Author information
Authors and Affiliations
Contributions
L Zhu, J Ye and M Sha were responsible for the overall design of the study and reviewing, revision of the manuscript; ZP Zheng and KY Li responsible for data acquisition, bioinformatics data processing and manuscript writing; Yang ZY and Wang XW were responsible for cell culture and molecular experimental validation; HM Lu and ZF Yin were responsible for animal rearing, modeling, and specimen extraction; C Shen and YB Zhang were responsible for the proofreading and statistical work of the experimental data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethical approval and consent to participate
The animal experiment of the study was approved by the Animal Welfare and Ethics Committee, Jiangsu Hanjiang Biotechnology Co., LTD (Ethics approval number: HJSW-23031301).
Consent for publication
Not applicable.
Additional information
Responsible Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, Z., Li, K., Yang, Z. et al. Transcriptomic analysis reveals molecular characterization and immune landscape of PANoptosis-related genes in atherosclerosis. Inflamm. Res. (2024). https://doi.org/10.1007/s00011-024-01877-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00011-024-01877-6