Abstract
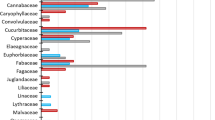
The emerging field of molecular scatology enables critical testing of food web theory. The non-invasive application of molecular tools allows for sequencing of prey DNA from predator fecal matter, evaluating diet breadth and foraging guild. While insectivorous bats are obscure foragers compared to most insectivorous birds, more is known about which arthropod species bats consume because molecular techniques have been optimized for mammalian systems, not avian physiology. Our research objective was to use molecular tools to detect arthropod prey in the fecal matter of an insectivorous avian predator. We used Western Bluebird (Sialia mexicana) as a model predator due to its generalist foraging strategy. We compared two fecal DNA extraction kits: (1) Qiagen’s DNA stool mini kits, used widely in dietary studies on bats and (2) Zymo’s Soil/Fecal DNA MiniPrep kits, not currently cited in the molecular scatology literature. We successfully extracted DNA only with the Zymo kit, amplified mitochondrial cytochrome oxidase c subunit I genes, sequenced, and identified the arthropod prey. A spiked PCR experiment showed evidence of possible inhibitors remaining in the Qiagen kit extractions. Overall, arthropod prey from seven different orders and five different classes were identified. We discuss the ecological implications of these data and suggest areas of future research applying molecular techniques to avian fecal matter. Consistent methodological advancement will enable molecular scatology to identify ecosystem services provided by insectivorous birds, develop ecological theory, and inform predator conservation efforts.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Beal F (1915) Food of the robins and bluebirds of the United States. U.S. Department of Agriculture Biological Survey Bulletin, vol 171, pp 151–175
Bohmann K, Monadjem A, Noer CL et al (2011) Molecular diet analysis of two African free-tailed bats (Molossidae) using high throughput sequencing. PLoS ONE 6:e21441. doi:10.1371/journal.pone.0021441
Bowles E, Schulte PM, Tollit DJ et al (2011) Proportion of prey consumed can be determined from faecal DNA using real-time PCR. Mol Ecol Resour 11:530–540
Carlisle JD, Holberton RL (2006) Relative efficiency of fecal versus regurgitated samples for assessing diet and the deleterious effects of a tartar emetic on migratory birds. J Field Ornithol 77:126–135. doi:10.1111/j.1557-9263.2006.00032.x
Clare EL, Fraser EE, Braid HE et al (2009) Species on the menu of a generalist predator, the eastern red bat (Lasiurus borealis): using a molecular approach to detect arthropod prey. Mol Ecol 18:2532–2542. doi:10.1111/j.1365-294X.2009.04184.x
Clare EL, Barber BR, Sweeney BW et al (2011) Eating local: influences of habitat on the diet of little brown bats (Myotis lucifugus). Mol Ecol 20:1772–1780. doi:10.1111/j.1365-294X.2011.05040.x
Corse E, Costedoat C, Chappaz R et al (2010) A PCR-based method for diet analysis in freshwater organisms using 18S rDNA barcoding on faeces. Mol Ecol Resour 10:96–108. doi:10.1111/j.1755-0998.2009.02795.x
Deagle BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from highly degraded samples—a case study on DNA in faeces. Front Zool 3:11
Deagle BE, Gales NJ, Evans K et al (2007) Studying seabird diet through genetic analysis of faeces: a case study on macaroni penguins (Eudyptes chrysolophus). PLoS ONE 2:e831. doi:10.1371/journal.pone.0000831
Deagle BE, Kirkwood R, Jarman SN (2009) Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces. Mol Ecol 18:2022–2038. doi:10.1111/j.1365-294X.2009.04158.x
Deagle BE, Chiaradia A, McInnes J, Jarman SN (2010) Pyrosequencing faecal DNA to determine diet of little penguins: is what goes in what comes out? Conserv Genet 11:2039–2048. doi:10.1007/s10592-010-0096-6
Folmer O, Black M, Hoeh W et al (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Guinan JA, Gowaty PA, Eltzroth EK (2008) Western Bluebird (Sialia mexicana). The birds of North America online. In: Poole A (ed) Ithaca: cornell lab of ornithology; retrieved from the birds of North America online: http://bna.birds.cornell.edu/bna/species/510. doi:10.2173/bna.510
Howard E, Heiser J (2004) What’s inside: anatomy and physiology. In: Podulka S, Rohrbaugh R, Bonney R (eds) Cornell lab of ornithology’s handbook of bird biology, chap 4, 2nd edn. Princeton University Press, Ithaca
Jarman SN, Gales NJ, Tierney M et al (2002) A DNA-based method for identification of krill species and its application to analysing the diet of marine vertebrate predators. Mol Ecol 11:2679–2690
King RA, Read DS, Traugott M, Symondson WOC (2008) Molecular analysis of predation: a review of best practice for DNA-based approaches. Mol Ecol 17:947–963. doi:10.1111/j.1365-294X.2007.03613.x
Leonard ML, Teather KL, Horn AG et al (1994) Provisioning in Western Bluebirds is not related to offspring sex. Behav Ecol 5:455–459. doi:10.1093/beheco/5.4.455
Levey DJ, Karasov WH (1992) Digestive modulation in a seasonal frugivore, the American Robin (Turdus migratorius). Am J Physiol 262:G711–G718
Levey DJ, Tewksbury JJ, Bolker BM (2008) Modelling long-distance seed dispersal in heterogeneous landscapes. J Ecol 96:599–608. doi:10.1111/j.1365-2745.2008.01401.x
Mellott RS, Woods PE (1993) An improved ligature technique for dietary sampling in nestling birds. J Field Ornithol 64:205–210
Moody DT (1970) A method for obtaining food samples from insectivorous birds. Auk 87:579
Murray DC, Bunce M, Cannell BL et al (2011) DNA-based faecal dietary analysis: a comparison of qPCR and high throughput sequencing approaches. PLoS ONE 6:e25776. doi:10.1371/journal.pone.0025776
Neuweiler G (2000) The biology of bats. Oxford University Press, New York
Newsome SD, Del Rio CM, Bearhop S, Phillips DL (2007) A niche for isotopic ecology. Front Ecol Environ 5:429–436. doi:10.1890/060150.01
Oehm J, Juen A, Nagiller K et al (2011) Molecular scatology: how to improve prey DNA detection success in avian faeces? Mol Ecol Resour 11:620–628
Pompanon F, Deagle BE, Symondson WOC et al (2011) Who is eating what: diet assessment using next generation sequencing. Mol Ecol. doi:10.1111/j.1365-294X.2011.05403.x
Razgour O, Clare EL, Zeale MRK et al (2011) High-throughput sequencing offers insight into mechanisms of resource partitioning in cryptic bat species. Ecol Evol 1:556–570. doi:10.1002/ece3.49
Scupham A, Jones J, Wesley I (2007) Comparison of DNA extraction methods for analysis of turkey cecal microbiota. J Appl Microbiol 102:401–409
Smith MA, Eveleigh ES, McCann KS et al (2011) Barcoding a quantified food web: crypsis, concepts, ecology and hypotheses. PLoS ONE 6:e14424. doi:10.1371/journal.pone.0014424
Symondson WOC (2002) Molecular identification of prey in predator diets. Mol Ecol 11:627–641
Valentini A, Miquel C, Nawaz MA et al (2009) New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Resour 9:51–60. doi:10.1111/j.1755-0998.2008.02352.x
Wilson JJ, Rougerie R, Schonfeld J et al (2011) When species matches are unavailable are DNA barcodes correctly assigned to higher taxa? an assessment using sphingid moths. BMC Ecol 11:18
Zeale MRK, Butlin RK, Barker GLA et al (2011) Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol Ecol Resour 11:236–244. doi:10.1111/j.1755-0998.2010.02920.x
Acknowledgments
National Science Foundation (DBI-1103583) and the University of California Agricultural Experiment Station provided funding for this project. We thank Ron Rosenbrand, Diane Kenworthy, Robert Burney, Joel Peterson, Bedrock Vineyards, and Spring Mountain Vineyards for access to bluebird nest boxes. We thank Marie Lefebvre for customizing a Perl script to obtain BLAST identifications for sequences. Four anonymous reviewers provided helpful comments on earlier versions of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jedlicka, J.A., Sharma, A.M. & Almeida, R.P.P. Molecular tools reveal diets of insectivorous birds from predator fecal matter. Conservation Genet Resour 5, 879–885 (2013). https://doi.org/10.1007/s12686-013-9900-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-013-9900-1