Abstract
GATA transcription factor family members have been found to play a critical role in the differentiation of many tissue types. For example, GATA-3 has been found to be highly correlated with estrogen receptor α (ER) expression and is emerging as one of the “master regulators” in breast ductal epithelial cell differentiation. Recently, we discovered another GATA family member highly prevalent in breast cancer called the trichorhinophalangeal syndrome-1 gene (TRPS-1). Using a quantitative immunohistochemistry (qIHC) approach, we found that TRPS-1 was significantly correlated with ER, PR, GATA-3, as well as HER2 expression. However, TRPS-1 was also found to be expressed in a high proportion of ER− ductal epithelial breast cancers (BCs), indicating that it may act as a ductal epithelial cell-specific transcription factor regulating cell fate at some point in the epithelial cell differentiation pathway. In keeping with this hypothesis, we found that TRPS-1 protein expression in BC above a certain threshold using qIHC correlated with markedly improved overall survival. Cox proportional hazards analysis found that both TRPS-1 and ER expression above critical threshold equally predicted for improved survival. Thus, TRPS-1 may be a powerful new positive prognostic marker in BC, and further IHC studies, as well as examination of its molecular function in ductal epithelial cell differentiation in the breast, are warranted. In this regard, data on the role of TRPS-1 in the differentiation of cells from mesenchymal precursors in other tissues, such as kidney metanephric mesenchymal cells, columnar chondrocytes, and osteoblasts, in mouse models may be useful. Indeed, these studies have found that TRPS-1 is a critical regulator of mesenchymal-to-epithelial cell transition. In the mammary gland, the restricted expression of TRPS-1 in human, mouse, and rat ductal epithelial cells suggests that it may also play a similar role during ductal luminal progenitor/stem cell differentiation. We present a model of TRPS-1 action in which it may act upstream of GATA-3 and ER on an earlier ductal epithelial progenitor cell or mammary stem cell during mammary gland development and also helps prevent reversion of ER+ BC cells back into mesenchymal-like cells. This model predicts that BCs with low or no TRPS-1 expression may inherently be much less differentiated and more aggressive tumors with less favorable prognosis.
Similar content being viewed by others
Introduction
Breast cancer (BC) is a heterogeneous disease. Most breast cancers are classified as invasive ductal carcinoma of the luminal type expressing the estrogen receptor α (ER) and dependent on estrogen for growth (about 70% of all BCs) [1]. ER+ BC is able to be effectively managed with anti-estrogen therapies [2, 3]. Overall, ER+ invasive ductal carcinoma is associated with a better prognosis than rarer HER2/neu+ERlo/− tumors and “basal type” tumors which are more apoptosis-resistant and have an unfavorable prognosis [4]. Although ER+ BC is a more manageable disease, a major problem is that 30% or more women eventually relapse while on anti-estrogen therapies [5, 6]. Another unsolved question is why estrogen sensitivity is lost in ER+ BC associated with the outgrowth of estrogen-independent variants associated with apoptosis resistance and higher rates of proliferation [6]. This estrogen-independent phenotype may be an inherent property of all ER+ BCs due to the presence of ER−, less differentiated cells present throughout the tumorigenic process that are selected for during anti-hormone signaling and other anti-tumor therapies. Thus, insight into this problem really becomes a question of understanding what regulates ductal epithelial cell differentiation and the acquisition and maintenance of an ER+, estrogen-sensitive ductal cell type intrinsic both during the development of ER+ ductal cancers as well as the normal mammary gland. Similarly, factors regulating the extent of epithelial cell differentiation in ER− and HER2-amplified BCs may play a similar role in regulating tumor aggressiveness and metastatic potential in a similar fashion. This suggests that a common set of factors may regulate the extent of cellular differentiation achieved before oncogenesis in all these BC subtypes. One way of gaining further insight into this problem is the identification of key gene expression pathways or “master regulators” switching on or off the major sets of genes regulating cell cycle activity and the expression genes involved in mesenchymal-to-epithelial cell transition (MET) in normal ductal cells and cells in invasive ductal BCs. One group of factors that has emerged to have such “master regulator” features controlling normal ductal epithelial and invasive ductal carcinoma cell differentiation is the GATA transcription factor family (e.g., GATA-3), acting in some cases in synergy with ER signaling. Identifying factors that disrupt GATA transcription factor function in early- and late-stage BC and how this is related to the loss of normal ER signaling may help elucidate how ER+ BC is initiated and how estrogen sensitivity is eventually lost. We recently reported that the trichorhinophalangeal syndrome-1 (TRPS-1) gene, a new GATA family transcription factor, is a ductal epithelial cell-specific gene expressed in normal breast and overexpressed to different extents in the majority of human BCs [7, 8]. We have recently reported that TRPS-1 is a marker for better prognosis when expressed over a critical threshold level using a new quantitative immunohistochemistry (qIHC) method applied to over 150 primary stage II/III BC cases of all subtypes [7]. This association with improved prognosis suggests that TRPS-1 is a tumor inhibitory gene. Moreover, its high prevalence in breast tumors, its association with ductal epithelial cells in normal breast, and its significant correlation with ER and GATA-3 suggest that it may be an essential regulator of luminal epithelial cell differentiation either independently or linked to the ER and GATA-3 signaling axis. This is supported by work in developmental biology that has found TRPS-1 to be a critical mediator of MET (9–13).
GATA Transcription Factor Family
The GATA transcription factor family is composed of six members (GATA-1 to GATA-6) having two prototypical zinc (Zn) fingers in their COOH-terminal region recognizing the consensus nucleotide sequence A/TGATAA/G in the promoter regions of target genes [14, 15]. The GATA transcription factor family was originally identified to be critical in mediating heart development and the differentiation of erythrocytes and lymphocytes (GATA-4, GATA-1, and GATA-3, respectively) [16, 17]. Recently, changes in GATA factor expression has been reported during tumorigenesis. Loss of GATA factor expression or inactivating mutations have been implicated in many tumors such as esophageal, prostate, and adrenocortical cancer (GATA-4 and GATA-5) [18–21]. GATA-3 also has an essential role in Th2 cell differentiation [22]. Recent studies have demonstrated that GATA-3 is the one of the most highly expressed transcription factors in the developing mammary gland during puberty and induces the expression of a set of genes required for normal mammary duct differentiation [23]. It is co-expressed with the ER and FOXA1 and other epithelial cell markers (e.g., cytokeratins and E-cadherin) in BC cell lines and during mammary gland development [24].
GATA-3 is Emerging as a “Master Regulator” in Breast Ductal Epithelial Cell Differentiation
In a microarray profiling study, GATA-3 was found to be one of the most highly enriched transcription factors in the mammary epithelium compared to stroma [23, 25]. Kouros-Mehr et al. and Asselin-Labat et al. have found that targeted disruption of GATA-3 expression in CD61+CD29loCD24+ luminal progenitor cells and ductal epithelial cells arising from terminal end bud (TEB) cells in the developing mammary gland in embryonic and adult mice leads to loss of normal cell cycle control and hyperplasia of undifferentiated cells associated with enhanced cell death [23, 26, 27]. Normal ductal architecture is completely disrupted together with the loss of expression of a number of differentiation-associated genes, including ER, FOXA1, cadherins, casein, amphiregulin, FOXP4, XBP1, and other factors associated with GATA-3 and ER signaling [23, 26]. FOXA1, a winged-helix transcription factor belonging to the forkhead family, is a chromatin modifier protein that facilitates chromatin opening, enhancing binding of ER to its target genes [28]. FOXA1 has been indicated to have a positive correlation with GATA-3, and further data have indicated that FOXA1 is the direct downstream target of GATA-3 transcriptional activity. It may act as a bridge between GATA-3 and ER [23]. Accumulating evidence suggests that GATA-3 cooperates with ER in driving mammary development and luminal ductal epithelial differentiation [27]. In a MMTV-PyMT mouse model of breast cancer, GATA-3 expression was lost early during tumor progression from adenoma to carcinoma [23]. This may have implications on how hormone sensitivity can be lost in ER+ BC. In contrast, GATA-3 overexpression has been found to reverse the metastatic activity of MDA-MB-231 BC cells (ER− and HER2− BC with low GATA-3 and low TRPS-1 expression) in a mouse xenograft model. This was correlated with a gain in E-cadherin and loss of vimentin, N-cadherin, and MMP9 [29].
Currently, it is unclear how GATA-3 gene expression itself is regulated in BC and during normal mammary gland development. Some evidence suggests that ER may drive GATA-3 and that GATA-3 also further drives ER expression in a reinforcing positive feedback loop [30]. This would explain the high ER and GATA-3 expression found in many hormone-dependent BC cases. However, other studies have found that estradiol activation of ER activity did not induce GATA-3 gene expression, making this still a controversial area that needs to be further studied [24]. Another possibility is that other transcriptional regulators acting further upstream in the system (i.e., on an earlier luminal progenitor cell) may also play a role in GATA-3 and ER expression, as well as genes encoding aromatases (e.g., CYP19) for estradiol synthesis by BC cells.
Although GATA-3 is emerging to be a critical regulator of ductal epithelial cell differentiation and loss of GATA-3 can be a driver of BC development, it is important to note that as a transcription factor operating in synergy with ER and driving FOXA1 and other ER-associated genes, GATA-3 also needs to be viewed as a driver of BC cell proliferation and survival. Thus, despite its role as a ductal epithelial cell differentiation factor, its presence does not prevent ductal carcinoma in situ (DCIS) and invasive ductal cancer development, but rather can also drive BC growth along with ER signaling. This “split personality” of GATA-3 is also reflected by the current controversy surrounding whether it is indeed an independent marker in BC.
GATA-3: A Possible Independent Prognostic Marker in BC, But Still Controversial
It has been widely accepted that ER+ BC patients have longer overall survival than those with ER− BC [31]. Since GATA-3 is one of the genes that are most highly correlated with ER expression, a number of studies have looked at its prognostic relevance in BC [24]. High expression of GATA-3 has been proposed to be a significant independent predictor of improved survival in several independent microarray datasets [24, 32]. Alternatively, low GATA-3 expression was suggested to be an independent indicator of poorer prognosis and high tumor grade [33]. However, these studies have been controversial, with more recent IHC studies on larger cohorts of BC patients finding that GATA-3 was not an independent prognostic marker [34–37]. In addition, another study found that in ER−/low, BC patients with a significant amount of GATA-3 expression had much poorer survival than patients without GATA-3 [38]. Mutations in GATA-3 (presumably knocking out its function) can also occur, but these are in <5% of BC cases [19]. In contrast, FOXA1 has been found to be an independent predictor of better prognosis in BC [25, 39]. Its expression is associated with the Luminal A BC subtype. The 30% or more of ER+ BC patients who do not respond to hormone therapy may be due to the absence of FOXA1 or lower levels of FOXA1 expression not meeting a critical threshold [37, 39].
The discovery of GATA-3 as a “master regulator” of ductal epithelial cell differentiation and ER sensitivity from a CD61+CD29loCD24+ luminal progenitor cell [26] suggests that perhaps other members of the GATA family may also operate during mammary gland development, perhaps even at an earlier stage. How committed luminal progenitors arise from earlier progenitors or mammary stem cells is still unknown and bears great relevance to BC. Thus, the discovery and characterization of additional GATA family members may yield further insight into this problem.
Overexpression of TRPS-1 in BC and its Potential Role in BC
Recently, we identified another GATA family transcription factor, called TRPS-1, from human BC cells after completing a large-scale gene expression profiling study of 73 BC RNA samples, including invasive ductal carcinomas, ductal carcinoma in situ, and metastatic lesions, in comparison to a large panel of 289 normal tissue RNA samples, including normal breast, to identify genes specifically overexpressed in BC [8]. We found that the TRPS-1 gene had the highest degree of prevalence in BC (83%), taking into consideration all major subtypes (ER+, triple-negative, and HER2-amplified). Table 1 shows the top-scoring genes overexpressed in BC in our original microarray screen [8]. The initial IHC analysis in this study also found that TRPS-1 is expressed in both ER+ and ER− BC and in normal breast ductal epithelial cells, albeit at lower levels than in BC cells [8]. However, the gene profiling studies have found on average a 3.8-fold increased gene expression in BC over normal breast tissue [8]. Interestingly, TRPS-1 was also found to be expressed in an androgen-dependent prostate cancer cell line (LNCaP) and tumors as a gene called GC79 by Chang et al. [40, 41]. In subsequent studies, the function of TRPS-1 in prostate cancer was explored. TRPS-1 was found overexpressed in androgen-dependent prostate cancer associated with the myc and the EIF3 gene at chromosome 8q24 [41, 42]. Data on whether any amplification of the TRPS-1 gene exists in BC and prostate cancer are not available. However, array CGH analysis of a large panel of BC samples indicated that TRPS-1 is not amplified (Gordon Mills, personal communication). At present, the genes regulated by TRPS-1 and its functional ramifications in BC are unclear, especially in context with the activity of GATA factors and ER signaling. Its overexpression and high prevalence, however, suggest that it has a critical role in regulating either BC initiation or progression.
IHC Analysis Reveals that TRPS-1 is a Potential Powerful Prognostic Marker in BC
Our initial IHC study found that 90% of the samples had 75% or more cells expressing TRPS-1 by IHC [8]. Due to this high prevalence of expression, we reasoned that traditional IHC scoring methods using subjective cutoffs for the degree of positive staining, as used with ER staining currently, may not be the best way to analyze differences in expression between patient samples. To address this problem, we developed a qIHC method to measure TRPS-1 protein expression that could be compared with tumor characteristic and patient clinical history [7]. Another reason why we developed a qIHC approach is that we have found that TRPS-1 RNA levels in many cases do not closely match protein levels found in BC cells and normal epithelial cells [7, 8]. In addition, we have found that some microarray platforms (e.g., Illumina™ bead-based system) do not accurately reflect TRPS-1 gene expression due to shortcomings in the probes used (Bao et al., unpublished observations). Because of this, TRPS-1 has been “missed” as a potentially important regulator in BC in many microarray-based studies. This underscores the fact that many critical genes in BC may be missed by researchers solely relying on microarray-based gene expression signatures aimed at uncovering important regulatory pathways in BC.
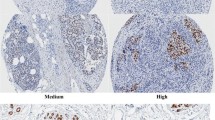
Using our qIHC approach, we have analyzed a set of 152 primary stage II/III invasive breast tumors. TRPS-1 was only expressed in the ductal epithelial cells and highly correlates ER and GATA-3. No staining of TRPS-1 was found in myoepithelial cells and basal cells and stromal tissue [7] similar to GATA-3, which is also restricted to the luminal epithelial cells and not expressed in myoepithelial cells [27]. We used a computerized image analysis system that combined the measured area of protein staining (called labeling index, LI) with the measured intensity of staining in each cell (called mean optical density, MOD) in a parameter called a “quick score” (QS), which was an overall quantitative measure of TRPS-1 expression in each tumor specimen. Quick score was calculated using the equation QS = LI × MOD/100 [7]. Using this approach, we found that TRPS-1 protein exhibited a considerable heterogeneity across the BC cases, as shown in representative stained specimens in Fig. 1a, b summarizing the distribution of TRPS-1 QS across all 152 stage II/III BC samples analyzed in this study, illustrating a wide distribution of expression. In some cases where adjacent normal ducts were visible in the specimens, we also determined the QS in the adjacent normal ducts in comparison to the BC cells. Overall, we found that the adjacent normal ducts always had detectable TRPS-1 expression, but this level of expression (QS) was lower than in the tumor. In ten such cases, the adjacent normal ducts had a median QS of 0.57, with the areas of invasive BC having a median QS of 12.58 [7]. DCIS had a median QS of 5.45, in between that of normal ducts and invasive cancer [7]. Another potentially important observation we made was that TRPS-1 expression in epithelial cells of the non-involved ducts seemed to parallel the levels seen in the adjacent tumor areas. In other words, patients with higher TRPS-1 in their tumor cells also had higher TRPS-1 in their adjacent normal ducts, and vice versa [7]. This suggests a provocative hypothesis that the molecular features in normal ducts and levels of TRPS-1 expression may be involved in regulating the pre-disposition to specific types of ductal cell cancers. In support of this, recent studies by Graham et al. [43, 44] found that the gene expression profiles of ER+ and ER− breast cancers were also detectable in histologically normal breast epithelium. They found that 25–53% of the BC-related genes in the ER+ and ER− BC samples studied were overlapping with their corresponding normal epithelium [43, 44].
TRPS-1 protein is found at different levels in the nuclei of ductal epithelial cells of BC from different patients using qIHC. a Staining and quantification of TRPS-1 expression using the QS parameter in representative patient BC samples illustrating the heterogeneity of TRPS-1 levels in BC. Magnification is at ×100. b Histogram showing the wide distribution of TRPS-1 QS across the stage II/III BC sample set studied from low to high QS
The heterogeneity in TRPS-1 expression in BC was further studied in a series of BC cell lines in vitro using real-time quantitative RT-PCR and immunoblot analysis, where TRPS-1 was found to be expressed at different levels across this panel of BC cell lines, with ER+ cell lines (such as T47D, CAMA-1, ZR75-1) tending to have a higher expression (Bao et al., unpublished observations). However, a number of ER− cell lines (MDA-MB-453 and SKBR3) also have high levels of TRPS-1 expression, suggesting a functional role in ER− BC cells, as suggested by our initial IHC studies that found TRPS-1 expression both in ER+ and ER− BC samples [8].
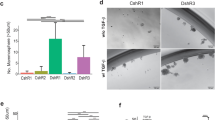
Kaplan–Meier survival analysis on the 152 stage II/III BC patients found a significantly improved survival of patients having a higher TRPS-1 QS versus lower TRPS-1 QS (Fig. 2a, left), similar to the survival curves generated when a critical ER expression threshold (QS ≥ 5 versus QS ≥ 5) was applied using this same qIHC approach (Fig. 2a, right). Cox proportional hazards analysis also showed that TRPS-1 could act as an equally predictive marker of improved survival as ER. As shown in Fig. 2b, TRPS-1 QS ≥ 4 was significantly associated with better survival, with a hazard ratio (HR) of 0.5 (p = 0.019), similar to ER QS ≥ 5 (HR = 0.48, p = 0.017). Interestingly, when we used for ER and GATA-3 either LI ≥5% or ≥10% (commonly used by most pathology labs), neither ER nor GATA-3 was associated with better or worse survival in a statistically significant manner (Fig. 2b) [7]. Thus, our data indicate that quantitative measurement of TRPS-1 expression can be a new prognostic test in BC with higher expression, a predictor of better survival. In addition, our data suggest that further qIHC studies on TRPS-1 are warranted using larger patient sample sets and in specific BC subtypes, where additional parameters such as relapse-free survival and time to relapse after chemotherapy or during anti-hormone therapy can be evaluated.
Quantitative IHC reveals that TRPS-1 may be a marker for improved survival in BC. a Kaplan–Meier survival analysis comparing the potential value of TRPS-1 as a marker for improved prognosis to that ER in a cohort of stage II/III BC patients (n = 152). Patients with TRPS-1 QS ≥ 4 had a significantly improved survival versus patients with TRPS-1 QS < 4 (left graph). ER expression was similarly associated with improved outcome with ER QS ≥ 5 group having a significantly longer overall survival than the QS < 5 group (right graph). The p values (p = 0.0165 for TRPS-1; p = 0.006 for ER), calculated using log rank analysis, are shown in each case. b Cox proportional hazards analysis shows TRPS-1 QS ≥ 4 to be significantly associated with better survival with a HR of 0.5 (p = 0.019), similar to ER QS ≥ 5 (HR = 0.48, p = 0.017). Notably, when the labeling index (LI) or percentage positive tumor cells in the samples were scored, ER and GATA-3 LI (≥5% or ≥10%) were not predictive (p ≥ 0.05)
TRPS-1 is Highly Conserved During Evolution and Expressed Only in Ductal Epithelial Cells of Human, Mouse, and Rat Mammary Glands
TRPS-1 has been classified as a new member of the GATA transcription factor with both similarities and key differences to the other GATA family members (GATA-1 through GATA-6). The human GATA genes have six exons and five introns with two homologous DNA-binding C4-type Zn finger domains encoded by exons IV and V separately, both similar in size. These Zn fingers bind a consensus sequence A/TGATAA/G in the promoter regions of target genes. Figure 3 shows the structure of the TRPS-1 protein. The protein has 1,281–1,294 amino acids in length with a molecular weight of approximately 141,521 Da. An alternative ATG start site, 39 bp upstream, may yield a 1,294-amino acid protein [45]. At present, however, the 1,281-amino acid version of TRPS-1 is considered as the full-length mature version expressed in mammalian cells [45]. TRPS-1 has seven C2H2-type Zn finger domains, a cysteine-rich region, a single GATA C4 -type Zn finger domain, and two C2H2-type IKAROS-like Zn finger domains [42]. The GATA domain of TRPS-1 has 77% homology with the C-terminal GATA domain of the superfamily of GATA transcription factors and has been shown to bind to the A/TGATAA/G consensus sequence [40, 46]. Thus, TRPS-1 is a new member of the GATA family, but is unusual in that it has only 1 GATA domain, unlike 2 GATA domains found in GATA 1 through 6 [46]. Functional analysis of TRPS-1 indicates that TRPS-1 plays a key role during embryonic development and as a modulator of cell differentiation and survival [10, 12, 47].
The TRPS-1 gene is highly conserved with 93% or higher amino acid sequence similarity between Xenopus, mouse, rat, and human [45, 46, 48, 49]. We have found that TRPS-1 is overexpressed in estradiol-induced BC in female ACI rats, an excellent animal model of human BC development [41, 48, 50–53]. This model uses a sub-panicular implantation of an estradiol pellet leading to a chronic moderate elevation of systemic estradiol levels in rat [41, 52]. This results in a spectrum of histological changes very similar to human BC, with the presence of usual hyperplasia, atypical ductal-associated hyperplasia, ductal carcinoma in situ, and invasive ductal carcinoma that follows in a sequence of events over a 5- to 6-month period of time [41, 48, 50–53]. Our anti-human TRPS-1 monoclonal antibodies (8D11 and 8A1) generated in our laboratory cross-reacts with both rat and mouse TRPS-1 [8]. Figure 4a shows the restricted expression of TRPS-1 in the ductal epithelial cells in normal breast of human, mouse, and the ACI rat by IHC using the 8D11 monoclonal antibody. Epitope mapping studies with this antibody suggests that it recognizes an N-terminal epitope that has 96% similarity between rat and human (Fig. 4b). Thus, the ACI rat estradiol-induced BC model would be a valuable tool to study the molecular role of TRPS-1 in invasive ER+ BC, DCIS, or other earlier stages, such as atypical ductal hyperplasia. Figure 4c shows TRPS-1 expression during all stages of ACI rat breast cancer progression.
Expression of TRPS-1 protein in human, mouse, and rat mammary glands and ductal breast tumors. a TRPS-1 is expressed in the normal ductal epithelial cells of the mammary gland of human, FVB normal mouse, and ACI rat. Magnification is at ×200. b The sequence of TRPS-1 at the amino terminus between rat and human shows high similarity. c TRPS-1 is expressed during all stages of 17β-estradiol induced (E2) ACI rat breast cancer progression from normal to hyperplasia (2 months of E2 treatment), focal dysplasia (red arrow, 3 months of E2 treatment), DCIS (4 months of E2 treatment), and development of invasive ductal tumors (6 months of E2 treatment). Magnification was at ×100. Tissue samples from ACI rats at different stages of tumor progression in response to E2 treatment were kindly provided by Dr. Jonathan Li and Dr. Sara Li (University of Kansas Medical Center, Kansas City, KS)
Transcriptional Function of TRPS-1
TRPS-1 was initially discovered as the gene associated with three rare autosomal genetic disorders called trichorhinophalangeal (TRP) syndromes caused by loss of heterozygosity at the q24 locus on chromosome 8 [45]. TRP syndrome patients have craniofacial and skeletal malformations as well as sparse hair due to the abnormal differentiation and incorrect patterning of cartilage, bone, and hair follicles during fetal and postnatal development [45]. The role of TRPS-1 in mammary gland development has not been addressed as of yet. TRP syndromes of different degrees of pathology are also associated with point mutations in one or both GATA zinc finger domains, or in the IKAROS domain [54–59]. The role of TRPS-1 in regulating cartilage, bone, and hair growth has also been addressed using targeted knockout models in mice that have TRP syndrome-like manifestations. TRPS-1−/− knockout mice die during embryonic development [12] with defects in multiple organs resembling human TRP syndromes [45]. TRPS-1ΔGT homozygote mice with a loss of TRPS-1 DNA binding die of respiratory failure shortly after birth due to a defect in bone and cartilage malformation and chondrocyte differentiation [60]. Further studies have demonstrated that TRPS-1 is a key modulator of chondrocyte and bone cell (osteoblast) differentiation and survival due to GATA-type zinc finger having the capacity to bind GATA consensus sequences [13, 59].
The single GATA zinc finger domain in TRPS-1 initially suggested that it may have a unique transcriptional regulatory role apart from the other GATA family members. A number of studies have suggested that TRPS-1 is a transcriptional repressor specifically binding to GATA domains upstream of GATA-dependent gene and inhibiting transcriptional activation by other GATA factors [12, 46, 61, 62]. At present, there is no evidence that TRPS-1 mediates ER, or the gene expression of GATA-3 or other GATA factors, but rather part of its function may be on regulating gene expression driven by GATA factors [12, 45, 59, 63]. In CHO cells, induced TRPS-1 expression was found to inhibit GATA transcriptional activation using luciferase reporter gene constructs [47]. In prostate cancer cells (LNCaP), TRPS-1 induction was shown to inhibit GATA-induced PSA expression [63] and promote apoptosis after androgen withdrawal [40, 64]. Recently, a proteomic analysis of DU145 prostate cancer cells using 2D gel electrophoresis and mass spectrometry found that TRPS-1 overexpression could suppress the expression of several antioxidant proteins, including superoxide dismutase, protein disulfide isomerase A3 precursor, endoplasmin precursor, and annexin A2 [65]. Additionally, TRPS-1 has been suggested to modulate a number of mitochondria-associated proteins, glycolytic enzymes, and proteins involved in apoptosis [65].
Possible Role of TRPS-1 in Development and Differentiation
The emerging role of TRPS-1 as a marker for improved prognosis, its specific expression in ductal epithelial cells in normal breast across multiple species, and its increased expression during BC progression suggest that TRPS-1 is a suppressive gene in BC and that it may play a critical role in driving a more differentiated epithelial cell phenotype. The results of some recent studies on TRPS-1 knockout mice and on the differentiation of mesenchymal cells in vitro indicate that such a critical role for TRPS-1 in BC and normal breast physiology does indeed exist. A summary of these findings is given below.
One of the most striking studies recently has implicated TRPS-1 as a critical regulator of renal tubule epithelial cell differentiation [10]. During early renal development in the embryo, TRPS-1 was observed to be co-expressed with bone morphogenetic protein 7 (BMP-7), a molecule having a key role in the differentiation of metanephric mesenchymal cells to kidney tubule epithelial cells [10]. TRPS-1 was found to be the key target of BMP-7 in its role in facilitating MET in developing kidney nephrons via the activation of the p38MAPK signaling pathway [10]. Recently, bioinformatics analysis has pointed out that TRPS-1 is one of the genes among 22 transcription factors (along with GATA-3) identified to be enriched in TEBs and mature ducts of mammary glands [23], again suggesting that in addition to GATA-3, TRPS-1 may play a role in luminal progenitor cell differentiation in the mammary gland. TRPS-1 also seems to be associated with osteoblast differentiation. In this case, TRPS-1 was found to directly or indirectly bind the osteocalcin promoter in the presence or absence of Runx2, mediating osteocalcin transcription [59]. TRPS-1 has also been proposed to be a modulator involved in MET during chondrocyte differentiation. These studies have shown that TRPS-1 regulates columnar chondrocyte differentiation and proliferation via interacting with Indian hedgehog (Ihh)/Gli3 signaling [13]. Inhibition of TRPS-1 expression prevents the normal differentiation of columnar chondrocytes (mesenchymal cells) into hypertrophic chondrocytes required for mature cartilage and bone formation. In this study, TRPS-1 was found to directly interact with the transactivation Gli3 through one of its zinc finger domains in vitro and in vivo. TRPS-1 seems to orchestrate a whole series of changes along with other cofactors in this chondrocyte differentiation pathway. TRPS-1 has also been found to act downstream of the Gdf5 signaling pathway in the chondrogenic cell line ATDC5. Gdf5 signaling induced TRPS-1 expression which promoted the differentiation of ATDC5 cells and their sensitivity to apoptosis by down-modulation of bcl-2 [11].
Studies on mesenchymal cell differentiation and the role of TRPS-1 and associated factors in chondrocytes, osteoblasts, and hair follicle cells cited above pave the way for a concrete avenue of investigation for elucidating the possible role of TRPS-1 in mammary gland development and BC. Although earlier studies suggested that TRPS-1 was strictly a transcriptional repressor of GATA-induced genes, the positive role of TRPS-1 in driving cellular differentiation, in conjunction with Runx2 and Gli3, opens up the possibility that it can act as a transcriptional activator under certain conditions, perhaps as a scaffold mediating the interaction of complexes of other transcription factors. In addition, other studies finding that dynein light chain protein LCA8 and ring finger protein 4 (RNF4) bind TRPS-1 and inhibit its GATA transcriptional repressor function further argue for a revised model of TRPS-1 action as a positive activator of key genes during cellular differentiation [61, 66]. Future studies will need to carefully assess what genes TRPS-1 may regulate using global chip on ChIP analysis and assessment of the functional and phenotypic consequences of TRPS-1 knockdown in BC cells, or induction in TRPS-1 in TRPS-1-negative BC cells.
Model for the Possible Function of TRPS-1 in Normal Ductal Epithelial Cell Differentiation and in the Regulation of Ductal BC
Based on the newly emerging transcriptional regulatory role of TRPS-1 as both a positive regulator of genes associated with MET and negative regulator of gene transcription induced by GATA factors, and the association of TRPS-1 with improved survival in BC, a plausible model for the function of TRPS-1 in mammary gland development and BC cell differentiation can be formulated based on the assumption that it is a BC “suppressor” gene. This model is illustrated in Fig. 5. TRPS-1 may emerge to be a key positive regulator of normal breast ductal epithelial cell differentiation while also helping to prevent the reverse process in ductal epithelial BC cells by preventing epithelial-to-mesenchymal transition (EMT). Normal as well as epithelial BC cells undergoing the EMT process have recently been shown to bare many of the hallmarks of BC stem cells [67]. The prevalent expression of TRPS-1 in ductal epithelial cell tumors having a wide variety of phenotypes (ER+, ER−, and HER2-amplified) suggests that it may in fact act upstream of other GATA factors such as GATA-3 in mediating mammary duct development. These findings, together with its role in mediating MET in other tissues, suggests TRPS-1 functions upstream of ER and GATA-3 in an earlier progenitor or mammary stem cell. We predict that breast tumors arising from TRPS-1-negative or TRPS-1-low precursor cells may be intrinsically more stem-like and more aggressive.
Abbreviations
- BC:
-
Breast cancer
- DCIS:
-
Ductal carcinoma in situ
- ER:
-
Estrogen receptor alpha
- IHC:
-
Immunohistochemistry
- qIHC:
-
Quantitative immunohistochemistry
- LI:
-
Labeling index
- MOD:
-
Mean optical density
- QS:
-
Quick score
- PSA:
-
Prostate-specific antigen
- TRPS-1:
-
Trichorhinophalangeal syndrome 1
- TEBs:
-
Terminal end buds
- MET:
-
Mesenchymal-to-epithelial cell transition
- EMT:
-
Epithelial-to-mesenchymal cell transition
- HR:
-
Hazard ratio
- CGH:
-
Comparative genomic hybridization
References
Keen JC, Davidson NE (2003) The biology of breast carcinoma. Cancer 97:825–833
Van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536
Dubart A, Romeo PH, Vainchenker W, Dumenil D (1996) Constitutive expression of GATA-1 interferes with the cell-cycle regulation. Blood 87:3711–3721
Freedman OC, Verma S, Clemons MJ (2005) Using aromatase inhibitors in the neoadjuvant setting: evolution or revolution? Cancer Treat Rev 31:1–17
Gordon NH, Silverman P, Lasheen W, Meinert J, Siminoff LA (2007) Thirty-year follow-up of chemo/hormonal therapy in node-positive breast cancer. Breast Cancer Res Treat 102:301–312
Tlsty TD (2007) Luminal cells GATA have it. Nat Cell Biol 9:135–136
Chen JQ, Litton J, Xiao L, Zhang HZ, Warneke CL, Wu Y, Shen X, Wu S, Sahin A, Katz R et al (2010) Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer 1:21–33
Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J et al (2005) The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA 102:11005–11010
Fantauzzo KA, Bazzi H, Jahoda CA, Christiano AM (2008) Dynamic expression of the zinc-finger transcription factor Trps1 during hair follicle morphogenesis and cycling. Gene Expr Patterns 8:51–57
Gai Z, Zhou G, Itoh S, Morimoto Y, Tanishima H, Hatamura I, Uetani K, Ito M, Muragaki Y (2009) Trps1 functions downstream of Bmp7 in kidney development. J Am Soc Nephrol 20:2403–2411
Itoh S, Kanno S, Gai Z, Suemoto H, Kawakatsu M, Tanishima H, Morimoto Y, Nishioka K, Hatamura I, Yoshida M et al (2008) Trps1 plays a pivotal role downstream of Gdf5 signaling in promoting chondrogenesis and apoptosis of ATDC5 cells. Genes Cells 13:355–363
Suemoto H, Muragaki Y, Nishioka K, Sato M, Ooshima A, Itoh S, Hatamura I, Ozaki M, Braun A, Gustafsson E et al (2007) Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev Biol 312:572–581
Wuelling M, Kaiser FJ, Buelens LA, Braunholz D, Shivdasani RA, Depping R, Vortkamp A (2009) Trps1, a regulator of chondrocyte proliferation and differentiation, interacts with the activator form of Gli3. Dev Biol 328:40–53
Viger RS, Taniguchi H, Robert NM, Tremblay JJ (2004) Role of the GATA family of transcription factors in andrology. J Androl 25:441–452
Molkentin JD (2000) The zinc finger-containing transcription factors GATA-4, -5, and -6 ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem 275:38949–38952
Crispino JD (2005) GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol 16:137–147
Patient RK, McGhee JD (2002) The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev 12:416–422
Charpentier AH, Bednarek AK, Daniel RL, Hawkins KA, Laflin KJ, Gaddis S, MacLeod MC, Aldaz CM (2000) Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res 60:5977–5983
Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG et al (2004) Mutation of GATA3 in human breast tumors. Oncogene 23:7669–7678
Guo M, Akiyama Y, House MG, Hooker CM, Heath E, Gabrielson E, Yang SC, Han Y, Baylin SB, Herman JG et al (2004) Hypermethylation of the GATA genes in lung cancer. Clin Cancer Res 10:7917–7924
Cullen ME, Patient RK (1997) GATA-1 DNA binding activity is down-regulated in late S phase in erythroid cells. J Biol Chem 272:2464–2469
Ho IC, Pai SY (2007) GATA-3—not just for Th2 cells anymore. Cell Mol Immunol 4:15–29
Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z (2006) GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127:1041–1055
Hoch RV, Thompson DA, Baker RJ, Weigel RJ (1999) GATA-3 is expressed in association with estrogen receptor in breast cancer. Int J Cancer 84:122–128
Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP (2007) FOXA1: growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer 120:1013–1022
Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J et al (2007) Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9:201–209
Kouros-Mehr H, Kim JW, Bechis SK, Werb Z (2008) GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol 20:164–170
Williamson EA, Wolf I, O’Kelly J, Bose S, Tanosaki S, Koeffler HP (2006) BRCA1 and FOXA1 proteins coregulate the expression of the cell cycle-dependent kinase inhibitor p27(Kip1). Oncogene 25:1391–1399
Yan W, Cao QJ, Arenas RB, Bentley B, Shao R (2010) GATA3 inhibits breast cancer metastasis through the reversal of epithelial–mesenchymal transition. J Biol Chem 285:14042–14051
Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M (2007) Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 67:6477–6483
Knight WA 3rd, Osborne CK, Yochmowitz MG, McGuire WL (1980) Steroid hormone receptors in the management of human breast cancer. Ann Clin Res 12:202–207
Bertucci F, Houlgatte R, Benziane A, Granjeaud S, Adelaide J, Tagett R, Loriod B, Jacquemier J, Viens P, Jordan B et al (2000) Gene expression profiling of primary breast carcinomas using arrays of candidate genes. Hum Mol Genet 9:2981–2991
Jenssen TK, Kuo WP, Stokke T, Hovig E (2002) Associations between gene expressions in breast cancer and patient survival. Hum Genet 111:411–420
Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG (2005) Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res 65:11259–11264
Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM (2006) Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24:1656–1664
Voduc D, Cheang M, Nielsen T (2008) GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomark Prev 17:365–373
Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW et al (2009) Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res 11:R40
Ciocca V, Daskalakis C, Ciocca RM, Ruiz-Orrico A, Palazzo JP (2009) The significance of GATA3 expression in breast cancer: a 10-year follow-up study. Hum Pathol 40:489–495
Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S (2008) Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol 61:327–332
Chang GTG, Steenbeek M, Schippers E, Blok LJ, van Weerden WM, van Alewijk DCJG, Eussen BHJ, van Steenbrugge GJ, Brinkmann AO (2000) Characterization of a zinc-finger protein and its association with apoptosis in prostate cancer cells. J Natl Cancer Inst 92:1414–1421
Li JJ, Li SA (2003) Causation and prevention of solely estrogen-induced oncogenesis: similarities to human ductal breast cancer. Adv Exp Med Biol 532:195–207
Chang GT, van den Bemd GJ, Jhamai M, Brinkmann AO (2002) Structure and function of GC79/TRPS1, a novel androgen-repressible apoptosis gene. Apoptosis 7:13–21
Graham K, de las Morenas A, Tripathi A, King C, Kavanah M, Mendez J, Stone M, Slama J, Miller M, Antoine G et al (2010) Gene expression in histologically normal epithelium from breast cancer patients and from cancer-free prophylactic mastectomy patients shares a similar profile. Br J Cancer 102:1284–1293
Graham K, Ge X, de Las Morenas A, Tripathi A, Rosenberg CL (2011) Gene expression profiles of estrogen receptor-positive and estrogen receptor-negative breast cancers are detectable in histologically normal breast epithelium. Clin Cancer Res 17:236–246
Momeni P, Glockner G, Schmidt O, von Holtum D, Albrecht B, Gillessen-Kaesbach G, Hennekam R, Meinecke P, Zabel B, Rosenthal A et al (2000) Mutations in a new gene, encoding a zinc-finger protein, cause tricho-rhino-phalangeal syndrome type I. Nat Genet 24:71–74
Malik TH, Shoichet SA, Latham P, Kroll TG, Peters LL, Shivdasani RA (2001) Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. EMBO J 20:1715–1725
Malik TH, Von Stechow D, Bronson RT, Shivdasani RA (2002) Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho-rhino-phalangeal syndromes. Mol Cell Biol 22:8592–8600
Li JJ, Weroha SJ, Lingle WL, Papa D, Salisbury JL, Li SA (2004) Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc Natl Acad Sci USA 101:18123–18128
van Agthoven T, van Agthoven TL, Dekker A, Foekens JA, Dorssers LC (1994) Induction of estrogen independence of ZR-75-1 human breast cancer cells by epigenetic alterations. Mol Endocrinol 8:1474–1483
Li JJ, Papa D, Davis MF, Weroha SJ, Aldaz CM, El-Bayoumy K, Ballenger J, Tawfik O, Li SA (2002) Ploidy differences between hormone- and chemical carcinogen-induced rat mammary neoplasms: comparison to invasive human ductal breast cancer. Mol Carcinog 33:56–65
Li SA, Weroha SJ, Tawfik O, Li JJ (2002) Prevention of solely estrogen-induced mammary tumors in female ACI rats by tamoxifen: evidence for estrogen receptor mediation. J Endocrinol 175:297–305
Turan VK, Sanchez RI, Li JJ, Li SA, Reuhl KR, Thomas PE, Conney AH, Gallo MA, Kauffman FC, Mesia-Vela S (2004) The effects of steroidal estrogens in ACI rat mammary carcinogenesis: 17beta-estradiol, 2-hydroxyestradiol, 4-hydroxyestradiol, 16alpha-hydroxyestradiol, and 4-hydroxyestrone. J Endocrinol 183:91–99
Weroha SJ, Li SA, Tawfik O, Li JJ (2006) Overexpression of cyclins D1 and D3 during estrogen-induced breast oncogenesis in female ACI rats. Carcinogenesis 27:491–498
Al-Dosari MS, Alkuraya FS (2010) Trichorhinophalangeal syndrome: report of a novel familial TRPS1 mutation. Clin Dysmorphol 19:98–100
Chen LH, Ning CC, Chao SC (2010) Mutations in TRPS1 gene in trichorhinophalangeal syndrome type I in Asian patients. Br J Dermatol 163:416–419
Hatamura I, Kanauchi Y, Takahara M, Fujiwara M, Muragaki Y, Ooshima A, Ogino T (2001) A nonsense mutation in TRPS1 in a Japanese family with tricho-rhino-phalangeal syndrome type I. Clin Genet 59:366–367
Ludecke HJ, Schaper J, Meinecke P, Momeni P, Gross S, von Holtum D, Hirche H, Abramowicz MJ, Albrecht B, Apacik C et al (2001) Genotypic and phenotypic spectrum in tricho-rhino-phalangeal syndrome types I and III. Am J Hum Genet 68:81–91
Piccione M, Niceta M, Antona V, Di Fiore A, Cariola F, Gentile M, Corsello G (2009) Identification of two new mutations in TRPS 1 gene leading to the tricho-rhino-phalangeal syndrome type I and III. Am J Med Genet A 149A:1837–1841
Piscopo DM, Johansen EB, Derynck R (2009) Identification of the GATA factor TRPS1 as a repressor of the osteocalcin promoter. J Biol Chem 284:31690–31703
Napierala D, Sam K, Morello R, Zheng Q, Munivez E, Shivdasani RA, Lee B (2008) Uncoupling of chondrocyte differentiation and perichondrial mineralization underlies the skeletal dysplasia in tricho-rhino-phalangeal syndrome. Hum Mol Genet 17:2244–2254
Kaiser FJ, Tavassoli K, Van den Bemd GJ, Chang GT, Horsthemke B, Moroy T, Ludecke HJ (2003) Nuclear interaction of the dynein light chain LC8a with the TRPS1 transcription factor suppresses the transcriptional repression activity of TRPS1. Hum Mol Genet 12:1349–1358
Kanno S, Gui T, Itoh S, Gai Z, Sun Y, Oikawa K, Yoshida M, Muragaki Y (2011) Aberrant expression of the P2 promoter-specific transcript Runx1 in epiphyseal cartilage of Trps1-null mice. Exp Mol Pathol 90:143–148
van den Bemd GJ, Jhamai M, Brinkmann AO, Chang GT (2003) The atypical GATA protein TRPS1 represses androgen-induced prostate-specific antigen expression in LNCaP prostate cancer cells. Biochem Biophys Res Commun 312:578–584
Chang GT, Jhamai M, van Weerden WM, Jenster G, Brinkmann AO (2004) The TRPS1 transcription factor: androgenic regulation in prostate cancer and high expression in breast cancer. Endocr Relat Cancer 11:815–822
Chang GT, Gamble SC, Jhamai M, Wait R, Bevan CL, Brinkmann AO (2007) Proteomic analysis of proteins regulated by TRPS1 transcription factor in DU145 prostate cancer cells. Biochim Biophys Acta 1774:575–582
Kaiser FJ, Moroy T, Chang GT, Horsthemke B, Ludecke HJ (2003) The RING finger protein RNF4, a co-regulator of transcription, interacts with the TRPS1 transcription factor. J Biol Chem 278:38780–38785
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M et al (2008) The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715
Acknowledgments
This work was supported by a grant from the Cancer Vaccine Program of Sanofi Pasteur Canada to MD Anderson Cancer Center (LS2006-00015466AG). J.Q.C. is a Susan G. Komen Breast Cancer Foundation Postdoctoral Fellow. Support from NIH grants P50 CA116199 and P50 CA116199-02S1 is also appreciated. We also thank Drs. Jonathan J Li and Sara Antonia Li (Department of Pharmacology, Toxicology and Therapeutics, The University of Kansas Medical Center) for kindly providing us specimens and slides of ACI rat mammary glands.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jie Qing Chen and Yi Bao contributed equally.
Rights and permissions
About this article
Cite this article
Chen, J.Q., Bao, Y., Litton, J. et al. Expression and Relevance of TRPS-1: A New GATA Transcription Factor in Breast Cancer. HORM CANC 2, 132–143 (2011). https://doi.org/10.1007/s12672-011-0067-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12672-011-0067-5