Abstract
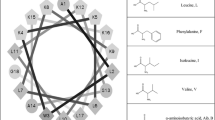
Antimicrobial peptides (AMPs) have been an area of great interest, due to the high selectivity of these molecules toward bacterial targets over host cells and the limited development of bacterial resistance to these molecules through evolution. The peptides are known to selectively bind to bacterial cell surfaces through electrostatic interactions, and subsequently, the peptides insert into the cell membrane and cause local disruptions of membrane integrity leading to cell death. Previous experiments showed that replacing the Leu residues in the AMP C18G with other naturally occurring hydrophobic residues resulted in side-chain-dependent activities. This work extends the investigation to non-natural hydrophobic amino acids and the effect on peptide activity. Minimal inhibitory concentration (MIC) results demonstrated that amino acid substitutions containing long flexible carbon chains maintained or increased antimicrobial activity compared to natural analogues. In solution, the peptide showed aggregation only with the most hydrophobic non-natural amino acid substitutions. Binding assays using Trp fluorescence confirm a binding preference for anionic lipids while quenching experiments demonstrated that the more hydrophobic peptides are more deeply buried in the anionic lipid bilayers compared to the zwitterionic bilayers. The most effective peptides at killing bacteria were also those which showed some level of disruption of bacterial membranes; however, one peptide sequence exhibited very strong activity and very low levels of red blood cell hemolysis, yielding a promising target for future development.







Similar content being viewed by others
References
Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. P T 40(4):277–283
Spellberg B, Gilbert DN (2014) The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis 59(Suppl 2):S71–S75. https://doi.org/10.1093/cid/ciu392
Rossiter SE, Fletcher MH, Wuest WM (2017) Natural products as platforms to overcome antibiotic resistance. Chem Rev 117(19):12415–12474. https://doi.org/10.1021/acs.chemrev.7b00283
Jakubczyk D, Dussart F (2020) Selected fungal natural products with antimicrobial properties. Molecules 25(4). https://doi.org/10.3390/molecules25040911
Kuroda K, Caputo GA (2013) Antimicrobial polymers as synthetic mimics of host-defense peptides. Wiley Interdiscip Rev Nanomed Nanobiotechnol 5(1):49–66. https://doi.org/10.1002/wnan.1199
Kuroki A, Kengmo Tchoupa A, Hartlieb M, Peltier R, Locock KES, Unnikrishnan M, Perrier S (2019) Targeting intracellular, multi-drug resistant Staphylococcus aureus with guanidinium polymers by elucidating the structure-activity relationship. Biomaterials 217:119249. https://doi.org/10.1016/j.biomaterials.2019.119249
Zhou Z, Ergene C, Lee JY, Shirley DJ, Carone BR, Caputo GA, Palermo EF (2019) Sequence and dispersity are determinants of photodynamic antibacterial activity exerted by peptidomimetic oligo(thiophene)s. ACS Appl Mater Interfaces 11(2):1896–1906. https://doi.org/10.1021/acsami.8b19098
Choi S, Isaacs A, Clements D, Liu D, Kim H, Scott RW, Winkler JD, DeGrado WF (2009) De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc Natl Acad Sci U S A 106(17):6968–6973. https://doi.org/10.1073/pnas.0811818106
Locock KES, Michl TD, Stevens N, Hayball JD, Vasilev K, Postma A, Griesser HJ, Meagher L, Haeussler M (2014) Antimicrobial polymethacrylates synthesized as mimics of tryptophan-rich cationic peptides. ACS Macro Lett 3(4):319–323. https://doi.org/10.1021/mz5001527
Takahashi H, Nadres ET, Kuroda K (2017) Cationic amphiphilic polymers with antimicrobial activity for oral care applications: eradication of S. mutans biofilm. Biomacromolecules 18(1):257–265. https://doi.org/10.1021/acs.biomac.6b01598
Goderecci SS, Kaiser E, Yanakas M, Norris Z, Scaturro J, Oszust R, Medina CD, Waechter F, Heon M, Krchnavek RR, Yu L, Lofland SE, Demarest RM, Caputo GA, Hettinger JD (2017) Silver oxide coatings with high silver-ion elution rates and characterization of bactericidal activity. Molecules 22(9). https://doi.org/10.3390/molecules22091487
Hajizadeh H, Peighambardoust SJ, Peighambardoust SH, Peressini D (2020) Physical, mechanical, and antibacterial characteristics of bio-nanocomposite films loaded with Ag-modified SiO2 and TiO2 nanoparticles. J Food Sci 85:1193–1202. https://doi.org/10.1111/1750-3841.15079
Vincent M, Duval RE, Hartemann P, Engels-Deutsch M (2018) Contact killing and antimicrobial properties of copper. J Appl Microbiol 124(5):1032–1046. https://doi.org/10.1111/jam.13681
O'Brien KT, Noto JG, Nichols-O'Neill L, Perez LJ (2015) Potent irreversible inhibitors of LasR quorum sensing in Pseudomonas aeruginosa. ACS Med Chem Lett 6(2):162–167. https://doi.org/10.1021/ml500459f
Sahariah P, Masson M (2017) Antimicrobial chitosan and chitosan derivatives: a review of the structure-activity relationship. Biomacromolecules 18(11):3846–3868. https://doi.org/10.1021/acs.biomac.7b01058
Cook K, Tarnawsky K, Swinton AJ, Yang DD, Senetra AS, Caputo GA, Carone BR, Vaden TD (2019) Correlating lipid membrane permeabilities of imidazolium ionic liquids with their cytotoxicities on yeast, bacterial, and mammalian cells. Biomolecules 9(6). https://doi.org/10.3390/biom9060251
Hanna SL, Huang JL, Swinton AJ, Caputo GA, Vaden TD (2017) Synergistic effects of polymyxin and ionic liquids on lipid vesicle membrane stability and aggregation. Biophys Chem 227:1–7. https://doi.org/10.1016/j.bpc.2017.05.002
Hice SA, Varona M, Brost A, Dai F, Anderson JL, Brehm-Stecher BF (2020) Magnetic ionic liquids: interactions with bacterial cells, behavior in aqueous suspension, and broader applications. Anal Bioanal Chem 412(8):1741–1755. https://doi.org/10.1007/s00216-020-02457-3
Sharma K, Aaghaz S, Shenmar K, Jain R (2018) Short antimicrobial peptides. Recent Pat Antiinfect Drug Discov 13(1):12–52. https://doi.org/10.2174/1574891X13666180628105928
Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ (2020) Antimicrobial host defence peptides: functions and clinical potential. Nat Rev Drug Discov 19:311–332. https://doi.org/10.1038/s41573-019-0058-8
Ponnappan N, Budagavi DP, Yadav BK, Chugh A (2015) Membrane-active peptides from marine organisms—antimicrobials, cell-penetrating peptides and peptide toxins: applications and prospects. Probiotics Antimicrob Proteins 7(1):75–89. https://doi.org/10.1007/s12602-014-9182-2
Ridgway Z, Picciano AL, Gosavi PM, Moroz YS, Angevine CE, Chavis AE, Reiner JE, Korendovych IV, Caputo GA (2015) Functional characterization of a melittin analog containing a non-natural tryptophan analog. Biopolymers 104(4):384–394. https://doi.org/10.1002/bip.22624
Kang HK, Kim C, Seo CH, Park Y (2017) The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J Microbiol 55(1):1–12. https://doi.org/10.1007/s12275-017-6452-1
Zasloff M (1987) Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A 84(15):5449–5453. https://doi.org/10.1073/pnas.84.15.5449
Ahmed TAE, Hammami R (2019) Recent insights into structure-function relationships of antimicrobial peptides. J Food Biochem 43(1):e12546. https://doi.org/10.1111/jfbc.12546
Wang G (2015) Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol 1268:43–66. https://doi.org/10.1007/978-1-4939-2285-7_3
Huang Y, Huang J, Chen Y (2010) Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell 1(2):143–152. https://doi.org/10.1007/s13238-010-0004-3
Eckert R (2011) Road to clinical efficacy: challenges and novel strategies for antimicrobial peptide development. Future Microbiol 6(6):635–651. https://doi.org/10.2217/fmb.11.27
Costa JR, Silva NC, Sarmento B, Pintado M (2017) Delivery systems for antimicrobial peptides and proteins: towards optimization of bioavailability and targeting. Curr Pharm Biotechnol 18(2):108–120
Kohn EM, Shirley DJ, Arotsky L, Picciano AM, Ridgway Z, Urban MW, Carone BR, Caputo GA (2018) Role of cationic side chains in the antimicrobial activity of C18G. Molecules 23(2). https://doi.org/10.3390/molecules23020329
Narancic T, Almahboub SA, O'Connor KE (2019) Unnatural amino acids: production and biotechnological potential. World J Microbiol Biotechnol 35(4):67. https://doi.org/10.1007/s11274-019-2642-9
Oliva R, Chino M, Pane K, Pistorio V, De Santis A, Pizzo E, D'Errico G, Pavone V, Lombardi A, Del Vecchio P, Notomista E, Nastri F, Petraccone L (2018) Exploring the role of unnatural amino acids in antimicrobial peptides. Sci Rep 8(1):8888. https://doi.org/10.1038/s41598-018-27231-5
Saint Jean KD, Henderson KD, Chrom CL, Abiuso LE, Renn LM, Caputo GA (2018) Effects of hydrophobic amino acid substitutions on antimicrobial peptide behavior. Probiotics Antimicrob Proteins 10(3):408–419. https://doi.org/10.1007/s12602-017-9345-z
Darveau RP, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, Maloney G (1992) Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest 90(2):447–455. https://doi.org/10.1172/JCI115880
Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI (2003) Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50(1):219–230. https://doi.org/10.1046/j.1365-2958.2003.03675.x
Park SY, Groisman EA (2014) Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol 91(1):135–144. https://doi.org/10.1111/mmi.12449
Hitchner MA, Santiago-Ortiz LE, Necelis MR, Shirley DJ, Palmer TJ, Tarnawsky KE, Vaden TD, Caputo GA (2019) Activity and characterization of a pH-sensitive antimicrobial peptide. Biochim Biophys Acta Biomembr 1861(10):182984. https://doi.org/10.1016/j.bbamem.2019.05.006
Caputo GA, London E (2019) Analyzing transmembrane protein and hydrophobic helix topography by dual fluorescence quenching. Methods Mol Biol 2003:351–368. https://doi.org/10.1007/978-1-4939-9512-7_15
Institute C-CaLS (2015) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard - tenth edition. CLSI document M07-A10
Burman LG, Nordström K, Boman HG (1968) Resistance of Escherichia coli to penicillins V. physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. J Bacteriol 96(2):438
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER suite: protein structure and function prediction. Nat Methods 12(1):7–8. https://doi.org/10.1038/nmeth.3213
Raghuraman H, Chattopadhyay A (2006) Effect of ionic strength on folding and aggregation of the hemolytic peptide melittin in solution. Biopolymers 83(2):111–121. https://doi.org/10.1002/bip.20536
Senetra AS, Necelis MR, Caputo GA (2020) Investigation of the structure-activity relationship in ponericin L1 from Neoponera goeldii. Pept Sci 112(3):e24162. https://doi.org/10.1002/pep2.24162
Chattopadhyay A, Haldar S (2014) Dynamic insight into protein structure utilizing red edge excitation shift. Acc Chem Res 47(1):12–19. https://doi.org/10.1021/ar400006z
Guha S, Rawat SS, Chattopadhyay A, Bhattacharyya B (1996) Tubulin conformation and dynamics: a red edge excitation shift study. Biochemistry 35(41):13426–13433. https://doi.org/10.1021/bi961251g
van Meer G, de Kroon AI (2011) Lipid map of the mammalian cell. J Cell Sci 124(Pt 1):5–8. https://doi.org/10.1242/jcs.071233
Kohn EM, Lee JY, Calabro A, Vaden TD, Caputo GA (2018) Heme dissociation from myoglobin in the presence of the zwitterionic detergent N,N-dimethyl-N-dodecylglycine betaine: effects of ionic liquids. Biomolecules 8(4). https://doi.org/10.3390/biom8040126
Palermo EF, Sovadinova I, Kuroda K (2009) Structural determinants of antimicrobial activity and biocompatibility in membrane-disrupting methacrylamide random copolymers. Biomacromolecules 10(11):3098–3107. https://doi.org/10.1021/bm900784x
Palermo EF, Kuroda K (2010) Structural determinants of antimicrobial activity in polymers which mimic host defense peptides. Appl Microbiol Biotechnol 87(5):1605–1615. https://doi.org/10.1007/s00253-010-2687-z
Thaker HD, Sgolastra F, Clements D, Scott RW, Tew GN (2011) Synthetic mimics of antimicrobial peptides from triaryl scaffolds. J Med Chem 54(7):2241–2254. https://doi.org/10.1021/jm101410t
Juretic D, Vukicevic D, Tossi A (2017) Tools for designing amphipathic helical antimicrobial peptides. Methods Mol Biol 1548:23–34. https://doi.org/10.1007/978-1-4939-6737-7_2
Jiang Z, Mant CT, Vasil M, Hodges RS (2018) Role of positively charged residues on the polar and non-polar faces of amphipathic alpha-helical antimicrobial peptides on specificity and selectivity for Gram-negative pathogens. Chem Biol Drug Des 91(1):75–92. https://doi.org/10.1111/cbdd.13058
Stark M, Liu LP, Deber CM (2002) Cationic hydrophobic peptides with antimicrobial activity. Antimicrob Agents Chemother 46(11):3585–3590. https://doi.org/10.1128/aac.46.11.3585-3590.2002
Avery CW, Palermo EF, McLaughlin A, Kuroda K, Chen Z (2011) Investigations of the interactions between synthetic antimicrobial polymers and substrate-supported lipid bilayers using sum frequency generation vibrational spectroscopy. Anal Chem 83(4):1342–1349. https://doi.org/10.1021/ac1025804
Torres MDT, Sothiselvam S, Lu TK, de la Fuente-Nunez C (2019) Peptide design principles for antimicrobial applications. J Mol Biol 431(18):3547–3567. https://doi.org/10.1016/j.jmb.2018.12.015
Zelezetsky I, Tossi A (2006) Alpha-helical antimicrobial peptides--using a sequence template to guide structure-activity relationship studies. Biochim Biophys Acta 1758(9):1436–1449. https://doi.org/10.1016/j.bbamem.2006.03.021
Lee DK, Brender JR, Sciacca MF, Krishnamoorthy J, Yu C, Ramamoorthy A (2013) Lipid composition-dependent membrane fragmentation and pore-forming mechanisms of membrane disruption by pexiganan (MSI-78). Biochemistry 52(19):3254–3263. https://doi.org/10.1021/bi400087n
Reshes G, Vanounou S, Fishov I, Feingold M (2008) Cell shape dynamics in Escherichia coli. Biophys J 94(1):251–264. https://doi.org/10.1529/biophysj.107.104398
Drin G, Antonny B (2010) Amphipathic helices and membrane curvature. FEBS Lett 584(9):1840–1847. https://doi.org/10.1016/j.febslet.2009.10.022
Fossati M, Goud B, Borgese N, Manneville JB (2014) An investigation of the effect of membrane curvature on transmembrane-domain dependent protein sorting in lipid bilayers. Cell Logist 4(2):e29087. https://doi.org/10.4161/cl.29087
Chen R, Mark AE (2011) The effect of membrane curvature on the conformation of antimicrobial peptides: implications for binding and the mechanism of action. Eur Biophys J 40(4):545–553. https://doi.org/10.1007/s00249-011-0677-4
Schmidt NW, Wong GC (2013) Antimicrobial peptides and induced membrane curvature: geometry, coordination chemistry, and molecular engineering. Curr Opin Solid State Mater Sci 17(4):151–163. https://doi.org/10.1016/j.cossms.2013.09.004
Matsuzaki K, Sugishita K, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM (1998) Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry 37(34):11856–11863. https://doi.org/10.1021/bi980539y
Acknowledgments
The authors would like to thank Dr. Dylan Klein for assistance with hemolysis assays and Mr. Calvin Caputo for assistance in synthesis of peptides.
Funding
This work was funded in part by NIH 1R15GM094330 to G.A.C.
Author information
Authors and Affiliations
Contributions
Caputo participated in research design; Hitchner, Necelis, and Shirley conducted the experiments and performed data analysis; Caputo, Necelis, and Hitchner contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1403 kb)
Rights and permissions
About this article
Cite this article
Hitchner, M.A., Necelis, M.R., Shirley, D. et al. Effect of Non-natural Hydrophobic Amino Acids on the Efficacy and Properties of the Antimicrobial Peptide C18G. Probiotics & Antimicro. Prot. 13, 527–541 (2021). https://doi.org/10.1007/s12602-020-09701-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-020-09701-3