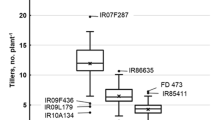
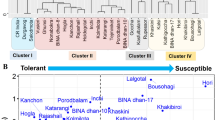
Abstract
The present study reports an unequivocal and improved protocol for efficient screening of salt tolerance at flowering stage in rice, which can aid phenotyping of population for subsequent identification of QTLs associated with salinity stress, particularly at reproductive stage. To validate the new method, the selection criteria, level and time of imposition of stress; plant growth medium were standardized using three rice genotypes. The setup was established with a piezometer placed in a perforated pot for continuous monitoring of soil EC and pH throughout the period of study. Further, fertilizer enriched soil was partially substituted by gravels for stabilization and maintaining the uniformity of soil EC in pots without hindering its buffering capacity. The protocol including modified medium (Soil:Stone, 4:1) at 8 dS m−1 salinity level was validated using seven different genotypes possessing differential salt sensitivity. Based on the important selection traits such as high stability index for plant yield, harvest index and number of grains/panicle and also high K+ concentration and low Na+– K+ ratio in flag leaf at grain filling stage were validated and employed in the evaluation of a mapping population in the modified screening medium. The method was found significantly efficient for easy maintenance of desired level of soil salinity and identification of genotypes tolerant to salinity at reproductive stage.





Similar content being viewed by others
Abbreviations
- PH:
-
Plant height (cm)
- PL:
-
Panicle length (cm)
- DAF:
-
Days to 50% flowering
- PN:
-
Panicle number/plant
- PY:
-
Plant yield (g)
- GRAIN:
-
Number of grains/plant
- DEG:
-
Spikelet degeneration (%)
- STE:
-
Spikelet sterility (%)
- FET:
-
Spikelet fertility (%)
- HI:
-
Harvest index
- SW:
-
Straw weight (g)
- EC:
-
Electrical conductivity (dS m−1)
- YSI:
-
Yield stability index
- K+ :
-
K+ concentration (µg/ml)
- Na+ :
-
Na+ concentration (µg/ml)
- Na–K:
-
Na+/K+ ratio
References
Ahmadizadeh M, Vispo NA, Calapit-Palao CDO, Pangaan ID, Vina CD, Singh RK (2016) Reproductive stage salinity tolerance in rice: a complex trait to phenotype. Ind J Plant Physiol. https://doi.org/10.1007/s40502-016-0268-6
Bhowmik SK, Islam MM, Emon RM, Begum SN, Siddika A, Sultana S (2007) Identification of salt tolerant rice cultivars via phenotypic and marker-assisted procedures. Pak J Biol Sci 10(24):4449–4454
Bouslama M, Schapaugh WT (1984) Stress tolerance in soybean. Part. I: evaluation of three screening techniques for heat and drought tolerance. Crop Sci 24:933–937
Chattopadhyay K, Nath D, Das G, Mohanta RL, Marndi BC, Singh DP, Sarkar RK, Singh ON (2013) Phenotyping and QTL-linked marker-based genotyping of rice lines with varying level of salt tolerance at flowering stage. Indian J Genet 73(4):434–437
Chattopadhyay K, Marndi BC, Sarkar RK, Singh ON (2017) Stability analysis of backcross population for salinity tolerance at reproductive stage in rice. Indian J Genet 77(1):51–58. https://doi.org/10.5958/0975-6906.2017.00007.4
Cho L, Yoon J, An G (2017) The control of flowering time by environmental factors. Plant J 90:708–719
Federer CA, Hornbeck JW (1985) The buffer capacity of forest soils in New England. Water Air Soil Pollut 26:163–173
Gregorio GB, Senadhira D, Mendoza RD (1997) Screening rice for salinity tolerance. IRRI discussion paper series no. 22, International Rice Research Institute, Manila (Philippines), pp 1-30
Hossain H, Rahman MA, Alam MS, Singh RK (2015) Mapping of quantitative trait loci associated with reproductive-stage salt tolerance in rice. J Agro Crop Sci. https://doi.org/10.1111/jac.12086
Iqbal SA, Islam MM, Hossain A, Malaker A (2015) DNA fingerprinting of rice lines for salinity tolerance at reproductive stage. Adv Crop Sci Technol. https://doi.org/10.4172/2329-8863.S1-006
Islam MR, Gregorio GB, Salam MA, Collard BCY, Singh RK, Hassan L (2012) Validation of SalTol linked markers and haplotype diversity on chromosome 1 of rice. Mol Plant Breed 3(10):103–114
Lutts S, Kinet JM, Bouharmont J (1995) Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot 46:1843–1852
Matoh T, Kairusmee P, Takahashi E (1986) Salt-induced damage to rice plants and alleviation effect of silicate. Soil Sci Plant Nutr 32(2):295–304
Mohammadi R, Vispo NAG, Amas JC, Sajise AG, Mamiit A, Gregorio GB (2011) Gravel-based hydroponic system: a new method for evaluating reproductive stage salinity tolerance in rice. Philippine J Crop Sci 36(1),http://agris.fao.org/aos/records/PH2013000609
Molla KA, Debnath AB, Ganie SA, Mondal TK (2015) Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice (Oryza sativa L.). BMC Plant Biol 15(1):1
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Rao PS, Mishra B, Gupta SR, Rathore A (2008) Reproductive stage tolerance to salinity and alkalinity stresses in rice genotypes. Plant Breed 127:256–261
Razzaque S, Haque T, Elias SM, Rahman MdS, Biswas S, Schwartz S, Ismail AM, Walia H, Juenger TE, Seraj ZI (2017) Reproductive stage physiological and transcriptional responses to salinity stress in reciprocal populations derived from tolerant (Horkuch) and susceptible (IR29) rice. Sci Rep 7:46138. https://doi.org/10.1038/srep46138
Ryu JY, Lee HJ, Seo PJ, Jung JH, Ahn JH, Park CM (2014) The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol Plant 7:377–387
Saha A, Sarkar RK, Yamagishi Y (1998) Effect of time of nitrogen application on spikelet differentiation and degeneration of rice. Bot Bull Acad Sin 39:119–123
Shereen A, Mumtaz S, Raza S, Khan MA, Solangi S (2005) Salinity effects on seedling growth and yield components of different inbred rice lines. Pak J Bot 37(1):131–139
Vijayan J, Senapati S, Ray S, Chakraborty K, Molla KA, Basak N, Pradhan B, Yeasmin L, Chattopadhyay K, Sarkar RK (2017) Transcriptomic and physiological studies identify cues for germination stage oxygen deficiency tolerance in rice. Environ Exp Bot 147:234–248
Vispo NA, Daep R, Mojares R, Pangaan ID, Babu AP, Singh RK (2015) A novel phenotyping technique for reproductive-stage salinity tolerance in rice. Procedia Environ Sci. https://doi.org/10.1016/j.proenv.2015.07.141
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. IRRI, Los Banos, pp 61–66
Yugandhar P, Veronica N, Panigrahy M, Rao DN, Subrahmanyam D, Voleti SR, Mangrauthia SK, Sharma RP, Sarla N (2017) Comparing hydroponics, sand, and soil medium to evaluate contrasting rice Nagina 22 mutants for tolerance to phosphorus deficiency. Crop Sci 57(4):2089–2097. https://doi.org/10.2135/cropsci2016.07.0594
Zaman SK, Chowdhury DAM, Bhyiyan NI (1997) The effect of salinity on germination, growth, yield and mineral composition of rice. Bangladesh J Agric Sci 24(1):103–109
Zeng L, Kwon TR, Xuan L, Wilson C, Grieve CM, Gregorio GB (2004) Genetic diversity analysis by microsatellite markers among rice genotypes with different adaptation to saline soils. Plant Sci 166:1258–1275
Acknowledgements
The financial support provided by the Director, ICAR-NRRI, Cuttack and the National Innovation on Climate Resilient Agriculture project (NICRA, ICAR), New Delhi, India are thankfully acknowledged.
Author information
Authors and Affiliations
Contributions
KC, AKN, BCM, AP and RKS performed the experiments. KC, AKN, KoC, and RKS discussed the results. KC, KoC and AKN wrote the article. All authors have approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chattopadhyay, K., Nayak, A.K., Marndi, B.C. et al. Novel screening protocol for precise phenotyping of salt-tolerance at reproductive stage in rice. Physiol Mol Biol Plants 24, 1047–1058 (2018). https://doi.org/10.1007/s12298-018-0591-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-018-0591-7