Abstract
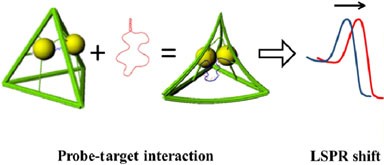
DNA origami is a promising technology for its reproducibility, flexibility, scalability and biocompatibility. Among the several potential applications, DNA origami has been proposed as a tool for drug delivery and as a contrast agent, since a conformational change upon specific target interaction may be used to release a drug or produce a physical signal, respectively. However, its conformation should be robust with respect to the properties of the medium in which either the recognition or the read-out take place, such as pressure, viscosity and any other unspecific interaction other than the desired target recognition. Here we report on the read-out robustness of a tetragonal DNA-origami/gold-nanoparticle hybrid structure able to change its configuration, which is transduced in a change of its plasmonic properties, upon interaction with a specific DNA target. We investigated its response when analyzed in three different media: aqueous solution, solid support and viscous gel. We show that, once a conformational variation is produced, it remains unaffected by the subsequent physical interactions with the environment.
Similar content being viewed by others
References
Ghosh, S. K.; Pal, T. Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: From theory to applications. Chem Rev. 2007, 107, 4797–4862.
Reinhard, B. M.; Siu, M.; Agarwal, H.; Alivisatos, A. P.; Liphardt, J. Calibration of dynamic molecular rulers based on plasmon coupling between gold nanoparticles. Nano Lett. 2005, 5, 2246–2252.
Prodan, E.; Radloff, C.; Halas, N. J.; Nordlander, P. A hybridization model for the plasmon response of complex nanostructures. Science2003, 302, 419–422.
Hill, R. T.; Mock, J. J.; Hucknall, A.; Wolter, S. D.; Jokerst, N. M.; Smith, D. R.; Chilkoti, A. Plasmon ruler with angstrom length resolution. ACS Nano2012, 6, 9237–9246.
Zhang, J.; Fu, Y.; Chowdhury, M. H.; Lakowicz, J. R. Metal-enhanced single-molecule fluorescence on silver particle monomer and dimer: Coupling effect between metal particles. Nano Lett. 2007, 7, 2101–2107.
Lim, D. K.; Jeon, K. S.; Kim, H. M.; Nam, J. M.; Suh, Y. D. Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection. Nat Mater. 2010, 9, 60–67.
Taminiau, T. H.; Stefani, F. D.; Segerink, F. B.; Van Hulst, N. F. Optical antennas direct single-molecule emission. Nat Photonics2008, 2, 234- 237.
Bek, A.; Jansen, R.; Ringler, M.; Mayilo, S.; Klar, T. A.; Feldmann, J. Fluorescence enhancement in hot spots of AFM-designed gold nanoparticle sandwiches. Nano Lett. 2008, 8, 485–490.
Ding, B. Q.; Deng, Z. T.; Yan, H.; Cabrini, S.; Zuckermann, R. N.; Bokor, J. Gold nanoparticle self-similar chain structure organized by DNA origami. J Am Chem Soc. 2010, 132, 3248–3249.
Zhou, C.; Duan, X. Y.; Liu, N. A plasmonic nanorod that walks on DNA origami. Nat Commun. 2015, 6, 8102.
Kuzyk, A.; Schreiber, R.; Fan, Z. Y.; Pardatscher, G.; Roller, E. M.; Högele, A.; Simmel, F. C.; Govorov, A. O.; Liedl, T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature2012, 483, 311–314.
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature2006, 440, 297–302.
Zanacchi, F. C.; Manzo, C.; Alvarez, A. S.; Derr, N. D.; Garcia-Parajo, M. F.; Lakadamyali, M. A DNA origami platform for quantifying protein copy number in super-resolution. Nat Methods2017, 14, 789–792.
Hudoba, M. W.; Luo, Y.; Zacharias, A.; Poirier, M. G.; Castro, C. E. Dynamic DNA origami device for measuring compressive depletion forces. ACS Nano2017, 11, 6566–6573.
Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem Rev. 2017, 11 7, 12584–12640.
Jiang, Q.; Song, C.; Nangreave, J.; Liu, X. W.; Lin, L.; Qiu, D. L.; Wang, Z. G.; Zou, G. Z.; Liang, X. J.; Yan, H. et al. DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc. 2012, 134, 13396- 13403.
Hemmig, E. A.; Fitzgerald, C.; Maffeo, C.; Hecker, L.; Ochmann, S. E.; Aksimentiev, A.; Tinnefeld, P.; Keyser, U. F. Optical voltage sensing using DNA origami. Nano Lett. 2018, 18, 1962–1971.
Marini, M.; Piantanida, L.; Musetti, R.; Bek, A.; Dong, M. D.; Besenbacher, F.; Lazzarino, M.; Firrao, G. A revertible, autonomous, self-assembled DNA-origami nanoactuator. Nano Lett. 2011, 11, 5449- 5454.
Torelli, E.; Marini, M.; Palmano, S.; Piantanida, L.; Polano, C.; Scarpellini, A.; Lazzarino, M.; Firrao, G. A DNA origami nanorobot controlled by nucleic acid hybridization. Small2014, 10, 2918–2926.
Prinz, J.; Schreiber, B.; Olejko, L.; Oertel, J.; Rackwitz, J.; Keller, A.; Bald, I. DNA origami substrates for highly sensitive surface-enhanced Raman scattering. J Phys Chem Lett. 2013, 4, 4140–4145.
Acuna, G. P.; Möller, F. M.; Holzmeister, P.; Beater, S.; Lalkens, B.; Tinnefeld, P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science2012, 338, 506–510.
Piantanida, L.; Naumenko, D.; Lazzarino, M. Highly efficient gold nanoparticle dimer formation via DNA hybridization. RSC Adv. 2014, 4, 15281–15287.
Sönnichsen, C.; Reinhard, B. M.; Liphardt, J.; Alivisatos, A. P. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nat Biotechnol. 2005, 23, 741–745.
Kuzyk, A.; Urban, M. J.; Idili, A.; Ricci, F.; Liu, N. Selective control of reconfigurable chiral plasmonic metamolecules. Sci Adv. 2017, 3, e1602803.
Zhou, C.; Xin, L.; Duan, X. Y.; Urban, M. J.; Liu, N. Dynamic plasmonic system that responds to thermal and aptamer-target regulations. Nano Lett. 2018, 18, 7395–7399.
Schreiber, R.; Luong, N.; Fan, Z. Y.; Kuzyk, A.; Nickels, P. C.; Zhang, T.; Smith, D. M.; Yurke, B.; Kuang, W.; Govorov, A. O. et al. Chiral plasmonic DNA nanostructures with switchable circular dichroism. Nat Commun. 2013, 4, 2948.
Piantanida, L.; Naumenko, D.; Torelli, E.; Marini, M.; Bauer, D. M.; Fruk, L.; Firrao, G.; Lazzarino, M. Plasmon resonance tuning using DNA origami actuation. Chem Commun. 2015, 51, 4789–4792.
Kim, D. N.; Kilchherr, F.; Dietz, H.; Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012, 40, 2862–2868.
Castro, C. E.; Kilchherr, F.; Kim, D. N.; Shiao, E. L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat Methods2011, 8, 221–229.
Masciotti, V.; Naumenko, D.; Lazzarino, M.; Piantanida, L. Tuning gold nanoparticles plasmonic properties by DNA nanotechnology. In DNA Nanotechnology: Methods and Protocols. Zuccheri, G., Ed.; Springer: New York, N Y, 2018; pp 279–297.
Dubochet, J.; Adrian, M.; Chang, J. J.; Homo, J. C.; Lepault, J.; McDowall, A. W.; Schultz, P. Cryo-electron microscopy of vitrified specimens. Quart Rev Biophys. 1988, 21, 129–228.
Glaeser, R. M. Retrospective: Radiation damage and its associated “information limitations”. J Struct Biol. 2008, 163, 271–276.
Lei, D. S.; Marras, A. E.; Liu, J. F.; Huang, C. M.; Zhou, L. F.; Castro, C. E.; Su, H. J.; Ren, G. Three-dimensional structural dynamics of DNA origami Bennett linkages using individual-particle electron tomography. Nat Commun. 2018, 9, 592.
Zhang, L.; Lei, D. S.; Smith, J. M.; Zhang, M.; Tong, H. M.; Zhang, X.; Lu, Z. Y.; Liu, J. K.; Alivisatos, A. P.; Ren, G. Three-dimensional structural dynamics and fluctuations of DNA-nanogold conjugates by individual-particle electron tomography. Nat Commun. 2016, 7, 11083.
Amenitsch, H.; Rappolt, M.; Kriechbaum, M.; Mio, H.; Laggner, P.; Bernstorff, S. First performance assessment of the small-angle X-ray scattering beamline at ELETTRA. J Synchrotron Radiat. 1998, 5, 506- 508.
Bernstorff, S.; Amenitsch, H.; Laggner, P. High-throughput asymmetric double-crystal monochromator of the SAXS beamline at ELETTRA. J Synchrotron Radiat. 1998, 5, 1215–1221.
Forget, A.; Pique, R. A.; Ahmadi, V.; Lüdeke, S.; Shastri, V. P. Mechanically tailored agarose hydrogels through molecular alloying with β-sheet polysaccharides. Macromol Rapid Commun. 2015, 36, 196–203.
Rüther, A.; Forget, A.; Roy, A.; Carballo, C.; Mießmer, F.; Dukor, R. K.; Nafie, L. A.; Johannessen, C.; Shastri, V. P.; Lüdeke, S. Unravelling a direct role for polysaccharide β-strands in the higher order structure of physical hydrogels. Angew Chem., Int Ed. 2017, 56, 4603–4607.
Mie, G. Beiträge zur Optik trüber Medien, speziell kolloidaler Metallösungen. Ann Phys. 1908, 330, 377–445.
García De Abajo, F. J. Multiple scattering of radiation in clusters of dielectrics. Phys Rev B1999, 60, 6086–6102.
Myroshnychenko, V.; Rodríguez-Fernández, J.; Pastoriza-Santos, I.; Funston, A. M.; Novo, C.; Mulvaney, P.; Liz-Marzán, L. M.; García De Abajo, F. J. Modelling the optical response of gold nanoparticles. Chem Soc Rev. 2008, 37, 1792–1805.
Walsh, A. S.; Yin, H. F.; Erben, C. M.; Wood, M. J. A.; Turberfield, A. J. DNA cage delivery to mammalian cells. ACS Nano2011, 5, 5427–5432.
Lee, H.; Lytton-Jean, A. K. R.; Chen, Y.; Love, K. T.; Park, A. I.; Karagiannis, E. D.; Sehgal, A.; Querbes, W.; Zurenko, C. S.; Jayaraman, M. et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012, 7, 389–393.
Acknowledgements
V. M. acknowledges financial support from MIUR (MIUR Giovani-Ambito “Salute dell’uomo”). Work at the Molecular Foundry, under the research project No. 3376, was supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We acknowledge the Facility of Nanofabrication (FNF) of IOM for the support in sample preparation, Simone Dal Zilio and Silvio Greco for help in data analysis and stimulating discussions. We acknowledge Prof. Giuseppe Firrao for valuable comments and inspiring ideas, the NanoInnovation laboratory (Elettra Sincrotrone) for suggestion provided for AFM analysis and the BioLab (Elettra Sincrotrone) for the use of lab and instrumentation.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Masciotti, V., Piantanida, L., Naumenko, D. et al. A DNA origami plasmonic sensor with environment-independent read-out. Nano Res. 12, 2900–2907 (2019). https://doi.org/10.1007/s12274-019-2535-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12274-019-2535-0