Abstract
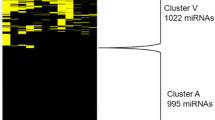
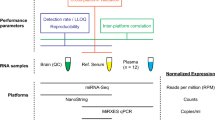
Microarray, RT-qPCR based arrays and next-generation-sequencing (NGS) are available high-throughput methods for miRNA profiling (miRNome). Analytical and biological performance of these methods were tested in identification of biologically relevant miRNAs in non-functioning pituitary adenomas (NFPA). miRNome of 4 normal pituitary (NP) and 8 NFPA samples was determined by these platforms and expression of 21 individual miRNAs was measured on 30 (20 NFPA and 10 NP) independent samples. Complex bioinformatics was used. 132 and 137 miRNAs were detected by all three platforms in NP and NFPA, respectively, of which 25 were differentially expressed (fold change > 2). The strongest correlation was observed between microarray and TaqMan-array, while the data obtained by NGS were the most discordant despite of various bioinformatics settings. As a technical validation we measured the expression of 21 selected miRNAs by individual RT-qPCR and we were able to validate 35.1%, 76.2% and 71.4% of the miRNAs revealed by SOLiD, TLDA and microarray result, respectively. We performed biological validation using an extended number of samples (20 NFPAs and 8 NPs). Technical and biological validation showed high correlation (p < 0.001; R = 0.96). Pathway and network analysis revealed several common pathways but no pathway showed the same activation score. Using the 25 platform-independent miRNAs developmental pathways were the top functional categories relevant for NFPA genesis. The difference among high-throughput platforms is of great importance and selection of screening method can influence experimental results. Validation by another platform is essential in order to avoid or to minimalize the platform specific errors.




Similar content being viewed by others
References
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858. https://doi.org/10.1126/science.1064921
Place RF, Li L-C, Pookot D et al (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105:1608–1613. https://doi.org/10.1073/pnas.0707594105
Ørom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30:460–471. https://doi.org/10.1016/j.molcel.2008.05.001
Zhang Z, Florez S, Gutierrez-Hartmann A et al (2010) MicroRNAs regulate pituitary development, and microRNA 26b specifically targets lymphoid enhancer factor 1 (Lef-1), which modulates pituitary transcription factor 1 (Pit-1) expression. J Biol Chem 285:34718–34728. https://doi.org/10.1074/jbc.M110.126441
Chen K, Rajewsky N (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 38:1452–1456. https://doi.org/10.1038/ng1910
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. https://doi.org/10.1016/j.cell.2004.12.035
Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro-Oncol 14(Suppl 5):v1–49. https://doi.org/10.1093/neuonc/nos218
Dworakowska D, Grossman AB (2009) The pathophysiology of pituitary adenomas. Best Pract Res Clin Endocrinol Metab 23:525–541. https://doi.org/10.1016/j.beem.2009.05.004
Mayson SE, Snyder PJ (2014) Silent (clinically nonfunctioning) pituitary adenomas. J Neuro-Oncol 117:429–436. https://doi.org/10.1007/s11060-014-1425-2
Sivapragasam M, Rotondo F, Lloyd RV et al (2011) MicroRNAs in the human pituitary. Endocr Pathol 22:134–143. https://doi.org/10.1007/s12022-011-9167-6
Li X-H, Wang EL, Zhou H-M et al (2014) MicroRNAs in Human Pituitary Adenomas. Int J Endocrinol 2014:435171. https://doi.org/10.1155/2014/435171
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369. https://doi.org/10.1038/nrg3198
Mestdagh P, Hartmann N, Baeriswyl L et al (2014) Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11:809–815. https://doi.org/10.1038/nmeth.3014
Git A, Dvinge H, Salmon-Divon M et al (2010) Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA N Y N 16:991–1006. https://doi.org/10.1261/rna.1947110
Butz H, Likó I, Czirják S et al (2010) Down-regulation of Wee1 kinase by a specific subset of microRNA in human sporadic pituitary adenomas. J Clin Endocrinol Metab 95:E181–E191. https://doi.org/10.1210/jc.2010-0581
Butz H, Németh K, Czenke D et al (2016) Systematic Investigation of Expression of G2/M Transition Genes Reveals CDC25 Alteration in Nonfunctioning Pituitary Adenomas. Pathol Oncol Res POR. https://doi.org/10.1007/s12253-016-0163-5
Thompson IR, Chand AN, King PJ et al (2012) Expression of guanylyl cyclase-B (GC-B/NPR2) receptors in normal human fetal pituitaries and human pituitary adenomas implicates a role for C-type natriuretic peptide. Endocr Relat Cancer 19:497–508. https://doi.org/10.1530/ERC-12-0129
Trivellin G, Butz H, Delhove J et al (2012) MicroRNA miR-107 is overexpressed in pituitary adenomas and inhibits the expression of aryl hydrocarbon receptor-interacting protein in vitro. Am J Physiol Endocrinol Metab 303:E708–E719. https://doi.org/10.1152/ajpendo.00546.2011
Harmati M, Tarnai Z, Decsi G et al (2016) Stressors alter intercellular communication and exosome profile of nasopharyngeal carcinoma cells. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. https://doi.org/10.1111/jop.12486
Butz H, Likó I, Czirják S et al (2011) MicroRNA profile indicates downregulation of the TGFβ pathway in sporadic non-functioning pituitary adenomas. Pituitary 14:112–124. https://doi.org/10.1007/s11102-010-0268-x
Butz H, Szabó PM, Nofech-Mozes R et al (2014) Integrative bioinformatics analysis reveals new prognostic biomarkers of clear cell renal cell carcinoma. Clin Chem 60:1314–1326. https://doi.org/10.1373/clinchem.2014.225854
Butz H, Szabó PM, Khella HWZ et al (2015) miRNA-target network reveals miR-124as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget 6:12543–12557. 10.18632/oncotarget.3815
Wang B, Howel P, Bruheim S et al (2011) Systematic evaluation of three microRNA profiling platforms: microarray, beads array, and quantitative real-time PCR array. PLoS One 6:e17167. https://doi.org/10.1371/journal.pone.0017167
Chevillet JR, Lee I, Briggs HA et al (2014) Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Mol Basel Switz 19:6080–6105. https://doi.org/10.3390/molecules19056080
Farr RJ, Januszewski AS, Joglekar MV et al (2015) A comparative analysis of high-throughput platforms for validation of a circulating microRNA signature in diabetic retinopathy. Sci Rep 5:10375. https://doi.org/10.1038/srep10375
Bottoni A, Zatelli MC, Ferracin M et al (2007) Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol 210:370–377. https://doi.org/10.1002/jcp.20832
Liang S, Chen L, Huang H, Zhi D (2013) The experimental study of miRNA in pituitary adenomas. Turk Neurosurg 23:721–727. https://doi.org/10.5137/1019-5149.JTN.7425-12.1
Chambers TJG, Giles A, Brabant G, Davis JRE (2013) Wnt signalling in pituitary development and tumorigenesis. Endocr Relat Cancer 20:R101–R111. https://doi.org/10.1530/ERC-13-0005
Elston MS, Gill AJ, Conaglen JV et al (2008) Wnt pathway inhibitors are strongly down-regulated in pituitary tumors. Endocrinology 149:1235–1242. https://doi.org/10.1210/en.2007-0542
Colli LM, Saggioro F, Serafini LN et al (2013) Components of the canonical and non-canonical Wnt pathways are not mis-expressed in pituitary tumors. PLoS One 8:e62424. https://doi.org/10.1371/journal.pone.0062424
Formosa R, Gruppetta M, Falzon S et al (2012) Expression and clinical significance of Wnt players and survivin in pituitary tumours. Endocr Pathol 23:123–131. https://doi.org/10.1007/s12022-012-9197-8
Lebrun J-J (2009) Activin, TGF-beta and menin in pituitary tumorigenesis. Adv Exp Med Biol 668:69–78
Ruebel KH, Leontovich AA, Tanizaki Y et al (2008) Effects of TGFbeta1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine 33:62–76. https://doi.org/10.1007/s12020-008-9060-3
Zhenye L, Chuzhong L, Youtu W et al (2014) The expression of TGF-β1, Smad3, phospho-Smad3 and Smad7 is correlated with the development and invasion of nonfunctioning pituitary adenomas. J Transl Med 12:71. https://doi.org/10.1186/1479-5876-12-71
Acknowledgements
This work has been funded by National Research, Development and Innovation Office – NKFIH PD116093 to Henriett Butz. Attila Patocs is a recipient of “Lendulet” grant from Hungarian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no potential conflicts of interest.
Electronic supplementary material
Supplemental Table 1
(DOCX 12 kb)
Supplemental Table 2
(DOCX 13 kb)
Supplemental Table 3
(DOCX 14 kb)
Supplemental Table 4
(DOCX 19 kb)
Supplementary Figure 1
Numbers of differentially expressed miRNAs in the same direction with a fold change cut-off 2 identified by different platforms (GIF 182 kb)
Supplementary Figure 2
a Pathway analysis of experimentally validated targets of differentially expressed miRNA lists obtained by different platforms b Comparison of most significant pathway activation z-scores of different platforms. Z-scores of pathways are presented where the scores showed the same direction at least two studies. Z-score infers the activation states of predicted pathway by investigating the activator or inhibitor function of the enriched genes in the particular pathway (GIF 43 kb)
Supplementary Figure 3
Pathway analysis of “platform independent” miRNAs (GIF 75 kb)
Supplementary Figure 4
Gene ontology analysis of “platform independent” miRNAs (GIF 183 kb)
Rights and permissions
About this article
Cite this article
Darvasi, O., Szabo, P.M., Nemeth, K. et al. Limitations of high throughput methods for miRNA expression profiles in non-functioning pituitary adenomas. Pathol. Oncol. Res. 25, 169–182 (2019). https://doi.org/10.1007/s12253-017-0330-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-017-0330-3