Abstract
Nearly complete backbone and side chain resonance assignments have been obtained for the third domain, residues M289–K400, of the envelope protein from the sylvatic strain (P72–1244) of the dengue 1 virus, containing mutations N336S and E370K, using double- and triple-resonance spectroscopy.
Similar content being viewed by others
Biological context
Dengue 1 virus (DEN1V) is a member of the family Flaviviridae, genus Flavivirus, which contains the dengue viruses (DEN 1–4), West Nile virus, Langat virus, Omsk hemorrhagic fever virus, and many others. Receptor binding and membrane fusion of flaviviruses occurs with the envelope (E) protein (Heinze and Allison 2003). The E protein ectodomain contains three domains, ED1, ED2, and ED3. The highly immunogenic ED3 is a putative receptor-binding domain and many neutralizing antibodies bind to ED3. Thus, ED3 is an attractive target for development of vaccines, anti-viral agents, and diagnostic antigens. Chemical shift assignments and/or NMR-based structures for the ED3 proteins of Japanese encephalitis (Wu et al. 2003), West Nile virus (Volk et al. 2004a, b), Omsk hemorrhagic fever virus (Volk et al. 2006a), Dengue 4 virus (Volk et al. 2006b, 2007b), Langat virus (Mukherjee et al. 2006), and yellow fever virus (Volk et al. 2007a) have previously been reported. In addition, crystallographic structures for the extra-membrane portion of the E protein have been reported for Tick-borne encephalitis virus (Rey et al. 1995), Dengue 2 virus (Modis et al. 2003), Dengue 3 virus (Modis et al. 2005), and West Nile virus (Kanai et al. 2006; Nybakken et al. 2006).
Methods and experiments
Uniformly 13C-, 15N-labeled recombinant DEN1-ED3 (sylvatic strain P72-1244) protein encompassing residues M289–K400, with mutations N366S and E370K, was expressed using the pET-15 vector (Novagen) in which the N-terminal His-tag sequence encoded in the plasmid was removed. Cells were lysed by sonication in native buffer, the crude cell debris was centrifuged and the supernatant was removed. The pellet was resuspended in denaturing buffer (6 M guanidine HCl) and the solution was passed through a size-exclusion column (G75 Sephadex®) to provide pure protein. Pooled fractions were dialyzed to remove the denaturant. A 3 kDa centricon (Amicon, Inc.) concentrator was used to concentrate the protein. The NMR sample contained 0.25 mM protein in 25 mM Tris–HCl (pH 6.5), 0.1% NaN3 and 25 mM NaCl in 90% H2O and 10% D2O.
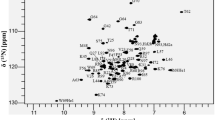
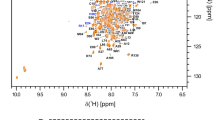
NMR experiments were performed at 25°C on Varian Inova 800 MHz (UTMB & Rice Univ.) or 600 MHz (Rice Univ.) spectrometers with triple-resonance coldprobes (Rice Univ.) and proton frequencies were referenced to internal DSS. The 13C and 15N dimensions were referenced indirectly using frequency ratios. Sequence-specific backbone assignments (Fig. 1) were obtained using the 2D 1H, 15N-HSQC, 3D HNCACB (Sattler et al. 1999), 3D CBCA(CO)NH, and 3D HNCO (Ikura et al. 1990; Bax and Grzesiek 1993) experiments. Non-aromatic side chain assignments were obtained using the TOCSY-[1H, 15N]-HSQC, H(C)CH-TOCSY (Clore and Gronenborn 1994), H(CCO)NH and C(CO)NH (Sattler et al. 1999) experiments. Aromatic proton assignments were obtained from the (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE experiments (Yamazaki et al. 1993). A NOESY-[1H, 15N]-HSQC experiment (Marion et al. 1989; Wüthrich 1986) provided several missing side chain assignments. The NMR spectra were processed in VNMRJ (Varian Inc.) or Felix2000 (Felix, Inc.) software.
Extent of assignments and data deposition
For the backbone atoms, 95% of the resonances have been assigned. The missing backbone atom resonances consist of one alpha carbon (N60), three carbonyl carbons (M1, N60 and P83) and 11 amide nitrogen atoms, corresponding to the six prolines, the N-terminal residues M1 and D2, and exposed residues Q28, G40 and N60.
The non-exchangeable proton side chain resonance assignments are nearly complete (99%), but many of the exchangeable proton/carbon pairs, such as the lysine HζCζ pairs and the HIS29 Hδ1/Nδ1 and Hε1/Nε1 pairs were not assigned. The aromatic carbon atoms were not assigned. The 1H, 13C, and 15N chemical shifts have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under the BMRB accession number 15782.
References
Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26:131–138. doi:10.1021/ar00028a001
Clore GM, Gronenborn AM (1994) Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol 239:249–363
Heinze FX, Allison S (2003) Flavivirus structure and membrane fusion. Adv Virus Res 59:63–97. doi:10.1016/S0065-3527(03)59003-0
Ikura M, Kay LE, Bax A (1990) A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Biochemistry 29:4659–4667. doi:10.1021/bi00471a022
Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E et al (2006) Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J Virol 80:11000–11008. doi:10.1128/JVI.01735-06
Marion D, Kay LE, Sparks SW, Torchia DA, Bax A (1989) Three dimensional heteronuclear NMR of 15N labeled proteins. J Am Chem Soc 111:1515–1517
Modis Y, Ogata S, Clements D, Harrison SC (2003) A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A 100:6986–6991. doi:10.1073/pnas.0832193100
Modis Y, Ogata S, Clements D, Harrison SC (2005) Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol 79:1223. doi:10.1128/JVI.79.2.1223-1231.2005
Mukherjee M, Dutta K, White MA, Cowburn D, Fox RO (2006) NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition. Protein Sci 15:1342–1355. doi:10.1110/ps.051844006
Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH (2006) Crystal structure of the West Nile virus envelope glycoprotein. J Virol 80:11467–11474. doi:10.1128/JVI.01125-06
Rey FA, Heinz FX, Mandl CW, Kunz C, Harrison SC (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:198–291. doi:10.1038/375291a0
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34:93–158. doi:10.1016/S0079-6565(98)00025-9
Volk DE, Beasley DWC, Kallick DA, Holbrook MR, Barrett ADT, Gorenstein DG (2004a) Solution structure and antibody binding studies of the envelope domain III from the New York strain of West Nile virus. J Biol Chem 279:38755–38761. doi:10.1074/jbc.M402385200
Volk DE, Chavez L, Beasley DWC, Barrett ADT, Holbrook MR, Gorenstein DG (2006a) Structure of the envelope protein domain III of Omsk hemorrhagic fever virus. Virology 351:188–195. doi:10.1016/j.virol.2006.03.030
Volk DE, Gandham SA, May FJ, Anderson A, Barrett ADT, Gorenstein DG (2007a) NMR assigments of the yellow fever virus envelope protein domain III. Biomol NMR Assign 1:49–50. doi:10.1007/s12104-007-9000-9
Volk DE, Kallick DA, Holbrook MR, Beasley DWC, Barrett ADT, Gorenstein DG (2004b) 1H, 13C, and 15N resonance assignments for the domain III of the West Nile virus envelope protein. J Biomol NMR 29:445–446. doi:10.1023/B:JNMR.0000032551.56061.68
Volk DE, Lee Y-C, Li X, Barrett ADT, Gorenstein DG (2006b) NMR assignments of the yellow fever virus envelope protein domain III. J Biomol NMR 36(Suppl 5):52. doi:10.1007/s10858-006-9048-3
Volk DE, Lee Y-C, Li X, Thiviyanathan V, Gromowski GD, Li L et al (2007b) Structure of the envelope protein domain III of dengue-4 virus. Virology 364:147–154. doi:10.1016/j.virol.2007.02.023
Wu K-P, Wu C-W, Tsao Y-F, Kuo T-W, Lou Y-C, Lin C-W et al (2003) Structural basis of a flavivirus recognized by its neutralizing antibody. J Biol Chem 278:46007–46013. doi:10.1074/jbc.M307776200
Wüthrich K (1986) NMR of protein and nucleic acids. Wiley, New York
Yamazaki T, Forman-Kay JD, Kay LE (1993) Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar coupling. Am Chem Soc 115:11054–11055. doi:10.1021/ja00076a099
Acknowledgments
This work was funded by the Pediatric Dengue Vaccine Initiative, the Centers for Disease Control (U90CCU618754), the National Institute for Allergic and Infectious Diseases (U01 AI054827), the Welch Foundation (H1296), and the State of Texas Advanced Technology Program (004952-0038-2003). We thank Sean Moran for assistance with the Rice University spectrometer whose carbon-enhanced HCN cold probe was obtained through the Strategic Partnership for Research in Nanotechnology Grant AFRL (AFOSR) FA9550-04-1-0328.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Volk, D.E., Anderson, K.M., Gandham, S.H.A. et al. NMR assignments of the sylvatic dengue 1 virus envelope protein domain III. Biomol NMR Assign 2, 155–157 (2008). https://doi.org/10.1007/s12104-008-9109-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-008-9109-5