Abstract
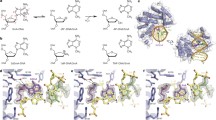
E. coli AlkB is a direct DNA/RNA repair protein that oxidatively reverses N1 alkylated purines and N3 alkylated pyrimidines to regular bases. Previous crystal structures have revealed N1-methyl adenine (1-meA) recognition by AlkB and a unique base flipping mechanism, but how the AlkB active site can accommodate bulky base adducts is largely unknown. Employing a previously developed chemical cross-linking technique, we crystallized AlkB with a duplex DNA containing a caged thymine base (cagedT). The structure revealed a flexible hairpin lid and a reorganized substrate recognition loop used by AlkB to accommodate cagedT. These observations demonstrate, at the molecular level, how bulky DNA adducts may be recognized and processed by AlkB.
Similar content being viewed by others
References
Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair, 2007, 6: 429–442
Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, 2002, 419: 174–178
Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 2002, 419: 178–182
Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, Slupphaug G, Otterlei M, Krokan HE. Human AlkB homolog 1 is a mitochondrial pro tein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem, 2008, 283: 25046–25056
Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A, 2002, 99: 16660–16665
Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature, 2003, 421: 859–863
Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O’connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem, 2005, 280: 39448–39459
Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, Mcdonough MA, Cunliffe S, Mcneill LA, Galvanovskis J, Rorsman P, Robins P, Prieur X, Coll AP, Ma M, Jovanovic Z, Farooqi IS, Sedgwick B, Barroso I, Lindahl T, Ponting CP, Ashcroft FM, O’rahilly S, Schofield CJ. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science, 2007, 318: 1469–1472
Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett, 2008, 582: 3313–3319
Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, Falnes PO, Krokan HE, Klungland A. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J, 2006, 25: 2189–2198
Yi C, Yang CG, He C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc Chem Res, 2009
Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage. Chem Rev, 2006, 106: 215–232
Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol, 2004, 5: 148–157
Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A, 2004, 101: 14051–14056
Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res, 2004, 32: 6260–6267
Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem, 2004, 279: 40470–40474
Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol, 2005, 12: 855–860
Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc, 2005, 127: 14594–14595
Dinglay S, Gold B, Sedgwick B. Repair in Escherichia coli AlkB mutants of abasic sites and 3-methyladenine residues in DNA. Mutat Res, 1998, 407: 109–116
Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature, 2006, 439: 879–884
Sundheim O, Vagbo CB, Bjoras M, Sousa MM, Talstad V, Aas PA, Drablos F, Krokan HE, Tainer JA, Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J, 2006, 25: 3389–3397
Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature, 2008, 452: 961–965
Mishina Y, He C. Probing the structure and function of the Escherichia coli DNA alkylation repair AlkB protein through chemical cross-linking. J Am Chem Soc, 2003, 125: 8730–8731
Mishina Y, Chen LX, He C. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli. J Am Chem Soc, 2004, 126: 16930–16936
Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. (2001, 57: 1373–1382). Acta Crystallogr Sec D-Bio Crystallogr, 2003, 59: 404–404
Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sec D-Bio Crystallogr, 2004, 60: 2126–2132
Bailey S. The Ccp4 suite: programs for protein crystallography. Acta Crystallogr Sec D-Bio Crystallogr, 1994, 50: 760–763
Delano W L D S, Palo Alto, Ca. 2002
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yi, C., He, C. AlkB recognition of a bulky DNA base adduct stabilized by chemical cross-linking. Sci. China Chem. 53, 86–90 (2010). https://doi.org/10.1007/s11426-010-0008-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-010-0008-0