Abstract
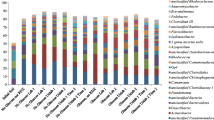
Vinyl chloride (VC) is a frequent groundwater contaminant and a known human carcinogen. Bioremediation is a potential cleanup strategy for contaminated sites; however, little is known about the bacteria responsible for aerobic VC degradation in mixed microbial communities. In attempts to address this knowledge gap, the microorganisms able to assimilate labeled carbon (13C) from VC within a mixed culture capable of rapid VC degradation (120 μmol in 7 days) were identified using stable isotope probing (SIP). For this, at two time points during VC degradation (days 3 and 7), DNA was extracted from replicate cultures initially supplied with labeled or unlabeled VC. The extracted DNA was ultracentrifuged, fractioned, and the fractions of greater buoyant density (heavy fractions, 1.758 to 1.780 g mL−1) were subject to high-throughput sequencing. Following this, specific primers were designed for the most abundant phylotypes in the heavy fractions. Then, quantitative PCR (qPCR) was used across the buoyant density gradient to confirm label uptake by these phylotypes. From qPCR and/or sequencing data, five phylotypes were found to be dominant in the heavy fractions, including Nocardioides (∼40 %), Sediminibacterium (∼25 %), Aquabacterium (∼17 %), Variovorax (∼6 %), and Pseudomonas (∼1 %). The abundance of two functional genes (etnC and etnE) associated with VC degradation was also investigated in the SIP fractions. Peak shifts of etnC and etnE gene abundance toward heavier fractions were observed, indicating uptake of 13C into the microorganisms harboring these genes. Analysis of the total microbial community indicated a significant dominance of Nocardioides over the other label-enriched phylotypes. Overall, the data indicate Nocardioides is primarily responsible for VC degradation in this mixed culture, with the other putative VC degraders generating a small growth benefit from VC degradation. The specific primers designed toward the putative VC degraders may be of use for investigating VC degradation potential at contaminated sites.







Similar content being viewed by others
References
Bradley PM (2003) History and ecology of chloroethene biodegradation: a review. Bioremed J 7:81–109
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108:4516–4522
Coleman NV, Spain JC (2003) Distribution of the coenzyme m pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl Environ Microb 69:6041–6046
Coleman NV, Mattes TE, Gossett JM, Spain JC (2002a) Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microb 68:6162–6171
Coleman NV, Mattes TE, Gossett JM, Spain JC (2002b) Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium. Appl Environ Microb 68:2726–2730
Cupples AM (2011) The use of nucleic acid based stable isotope probing to identify the microorganisms responsible for anaerobic benzene and toluene biodegradation. J Micro Meth 85:83–91
Cupples AM, Spormann AM, McCarty PL (2003) Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microb 69:953–959
Danko AS, Luo MZ, Bagwell CE, Brigmon RL, Freedman DL (2004) Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl Environ Microb 70:6092–6097
Davis J, Carpenter C (1990) Aerobic degradation of vinyl chloride in groundwater samples. Appl Environ Microb 56:3878–3880
Elango VK, Liggenstoffer AS, Fathepure BZ (2006) Biodegradation of vinyl chloride and cis-dichloroethene by a Ralstonia sp. strain TRW-1. Appl Micro Biotec 72:1270–1275
Fathepure BZ, Elango VK, Singh H, Bruner MA (2005) Bioaugmentation potential of a vinyl chloride-assimilating Mycobacterium sp., isolated from a chloroethene-contaminated aquifer. FEMS Micro Lett 248:227–234
Findlay M, Smoler DF, Fogel S, Mattes TE (2016) Aerobic vinyl chloride metabolism in groundwater microcosms by methanotrophic and etheneotrophic bacteria. Environ Sci Technol 50:3617–3625
Fullerton H, Rogers R, Freedman D, Zinder S (2014) Isolation of an aerobic vinyl chloride oxidizer from anaerobic groundwater. Biodegradation 25:893–901
Gossett JM (1987) Measurement of Henrys law constants for C1 and C2 chlorinated hydrocarbons. Environ Sci Technol 21:202–208
Guan XY, Liu F, Xie YX, Zhu LL, Han B (2013) Microbiota associated with the migration and transformation of chlorinated aliphatic hydrocarbons in groundwater. Environ Geochem Health 35:535–549
Hartmans S, Debont JAM (1992) Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl Environ Microb 58:1220–1226
Hartmans S, Debont JAM, Tramper J, Luyben K (1985) Bacterial degradation of vinyl chloride. Biotechnol Lett 7:383–388
Hatt JK, Loffler FE (2012) Quantitative real-time PCR (qPCR) detection chemistries affect enumeration of the Dehalococcoides 16S rRNA gene in groundwater. J Micro Meth 88:263–270
He J, Ritalahti KM, Yang K-L, Koenigsberg SS, Löffler FE (2003) Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65
Jin YO, Mattes TE (2008) Adaptation of aerobic, ethene-assimilating Mycobacterium strains to vinyl chloride as a growth substrate. Environ Sci Technol 42:4784–4789
Jin YO, Mattes TE (2010) A quantitative PCR assay for aerobic, vinyl chloride- and ethene-assimilating microorganisms in groundwater. Environ Sci Technol 44:9036–9041
Jin YO, Cheung S, Coleman NV, Mattes TE (2010) Association of missense mutations in epoxyalkane coenzyme M transferase with adaptation of Mycobacterium sp. strain JS623 to growth on vinyl chloride. Appl Environ Microb 76:3413–3419
Kanitkar Y, Stedtfeld RD, Steffan RJ, Hashsham SA, Cupples AM (2016) Development of loop mediated isothermal amplification (LAMP) for rapid detection and quantification of Dehalococcoides spp. biomarker genes in commercial reductive dechlorinating cultures KB-1 and SDC-9. Appl Environ Microb, In Press
Loffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Muller JA, Fullerton H, Zinder SH, Spormann AM (2013) Dehalococcoides mccartyi gen. nov., sp nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Micr 63:625–635
Mattes TE, Coleman NV, Spain JC, Gossett JM (2005) Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch Micro 183:95–106
Mattes TE, Alexander AK, Coleman NV (2010) Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Micro Rev 34:445–475
Paes F, Liu X, Mattes TE, Cupples AM (2015) Elucidating carbon uptake from vinyl chloride using stable isotope probing and Illumina sequencing. Appl Micro Biotec 99:7735–7743
Patterson BM, Aravena R, Davis GB, Furness AJ, Bastow TP, Bouchard D (2013) Multiple lines of evidence to demonstrate vinyl chloride aerobic biodegradation in the vadose zone, and factors controlling rates. J Contam Hydrol 153:69–77
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Gloeckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Radajewski S, Ineson P, Parekh NR, Murrell JC (2000) Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649
Ritalahti KM, Amos BK, Sung Y, Wu QZ, Koenigsberg SS, Loffler FE (2006) Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microb 72:2765–2774
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75:7537–7541
Stedtfeld RD, Stedtfeld TM, Kronlein M, Seyrig G, Steffan RJ, Cupples AM, Hashsham SA (2014) DNA extraction-free quantification of Dehalococcoides spp. in groundwater using a hand-held device. Environ Sci Technol 48:13855–13863
Sun WM, Cupples AM (2012) Diversity of five anaerobic toluene-degrading microbial communities investigated using stable isotope probing. Appl Environ Microb 78:972–980
Taylor AE, Dolan ME, Bottomley PJ, Semprini L (2007) Utilization of fluoroethene as a surrogate for aerobic vinyl chloride transformation. Environ Sci Technol 41:6378–6383
Verce MF, Ulrich RL, Freedman DL (2000) Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl Environ Microb 66:3535–3542
Acknowledgments
A collaborative NSF Grant (number 1233154) awarded to Timothy E. Mattes and Alison M. Cupples funded this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This work did not involve research with animals or humans.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Gerald Thouand
Rights and permissions
About this article
Cite this article
Wilson, F.P., Liu, X., Mattes, T.E. et al. Nocardioides, Sediminibacterium, Aquabacterium, Variovorax, and Pseudomonas linked to carbon uptake during aerobic vinyl chloride biodegradation. Environ Sci Pollut Res 23, 19062–19070 (2016). https://doi.org/10.1007/s11356-016-7099-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-016-7099-x