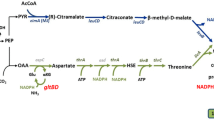
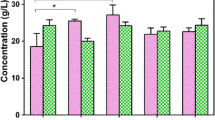
Abstract
Propionic acid (PA) is an important platform chemical used in the agriculture, food, and pharmaceutical industries. The biosynthesis of PA by propionibacteria has become an attractive alternative to traditional petrochemical processes owing to the environmentally friendly features of biorefinery. In a previous study, we significantly increased PA production in Propionibacterium acidipropionici by improving acid tolerance via genome shuffling. In this study, we undertook metabolomics analysis of parental P. acidipropionici and its genome-shuffled mutant to find the key metabolic nodes influencing PA production. In total, 142 intracellular metabolites were identified, of which those produced in amounts of greater than twofold difference between the two strains were further investigated with principal components analysis. The regulatory functions of key metabolites involved in the PA biosynthetic pathway were also forecast and analyzed according to their potential impact on metabolism. The results indicated that the amounts of metabolic intermediates of glycolysis, the Wood–Werkman cycle, and amino acid metabolism differed markedly between parental P. acidipropionici and its mutants. Based on the results of comparative metabolomics analysis, exogenous addition of key metabolites (precursors and amino acids) was performed to improve PA production. Under optimized conditions, 105 mM lactate, 20 mM fumarate, and 30 mM succinate were added to the culture of P. acidipropionici CGMCC 1.2230 in a 3-L anaerobic fermenter, and the PA titer increased from 23.1 ± 12 to 35.8 ± 1.0 g/L. This study revealed the key metabolic nodes of PA synthesis in P. acidipropionici through comparative metabolomics analysis, which may be helpful for the metabolic engineering of P. acidipropionici for improved PA production.





Similar content being viewed by others
References
Arbona, V., Manzi, M., Ollas, C. D., & Gómez-Cadenas, A. (2013). Metabolomics as a tool to investigate abiotic stress tolerance in plants. International Journal of Molecular Sciences, 14(3), 4885–4911.
Barding, G. A, Jr, Béni, S., Fukao, T., Bailey-Serres, J., & Larive, C. K. (2012). Comparison of GC-MS and NMR for metabolite profiling of rice subjected to submergence stress. Journal of Proteome Research, 12(2), 898–909.
Castillo, S., Gopalacharyulu, P., Yetukuri, L., & Orešič, M. (2011). Algorithms and tools for the preprocessing of LC-MS metabolomics data. Chemometrics and Intelligent Laboratory Systems, 108(1), 23–32.
Chen, F., Fen, X., Xu, H., Zhang, D., & Ouyang, P. (2012). Propionic acid production in a plant fibrous-bed bioreactor with immobilized Propionibacterium freudenreichii CCTCC M207015. Journal of Biotechnology, 164, 202–210.
Choi, H. K., Yoon, J. H., Kim, Y. S., & Kwon, D. Y. (2007). Metabolomic profiling of Cheonggukjang during fermentation by 1H NMR spectrometry and principal components analysis. Process Biochemistry, 42(2), 263–266.
Coral, J., Karp, S. G., de Souza Vandenberghe, L. P., Parada, J. L., Pandey, A., & Soccol, C. R. (2008). Batch fermentation model of propionic acid production by Propionibacterium acidipropionici in different carbon sources. Applied Biochemistry and Biotechnology, 151(2–3), 333–341.
Deborde, C., & Boyaval, P. (2000). Interactions between pyruvate and lactate metabolism in Propionibacterium freudenreichii subsp. shermanii: In vivo 13C nuclear magnetic resonance studies. Applied and Environmental Microbiology, 66(5), 2012–2020.
Dietmair, S., Hodson, M. P., Quek, L. E., Timmins, N. E., Chrysanthopoulos, P., Jacob, S. S., et al. (2012). Metabolite profiling of CHO cells with different growth characteristics. Biotechnology and Bioengineering, 109(6), 1404–1414.
Ding, M. Z., Zhou, X., & Yuan, Y. J. (2010). Metabolome profiling reveals adaptive evolution of Saccharomyces cerevisiae during repeated vacuum fermentations. Metabolomics, 6(1), 42–55.
Du, J., Zhou, J., Xue, J., Song, H., & Yuan, Y. (2012). Metabolomic profiling elucidates community dynamics of the Ketogulonicigenium vulgare-Bacillus megaterium consortium. Metabolomics, 8(5), 960–973.
Feng, X., Chen, F., Xu, H., Wu, B., Li, H., Li, S., et al. (2011). Green and economical production of propionic acid by Propionibacterium freudenreichii CCTCC M207015 in plant fibrous-bed bioreactor. Bioresource technology, 102(10), 6141–6146.
Guan, N., Liu, L., Shin, H. D., Chen, R. R., Zhang, J., Li, J., et al. (2013). Systems-level understanding how Propionibacterium acidipropionici respond to propionic acid stress at the microenvironment levels: Mechanism and application. Journal of Biotechnology, 167(1), 56–63.
Guan, N., Liu, L., Zhuge, X., Xu, Q., Li, J., Du, C., et al. (2012). Genome-shuffling improves acid tolerance of Propionibacterium acidipropionici and propionic acid production. Advances in Chemistry Research, 15, 143–152.
Hasunuma, T., Sanda, T., Yamada, R., Yoshimura, K., Ishii, J., & Kondo, A. (2011). Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microbial Cell Factories, 10, 2–14.
Jin, Z., & Yang, S. T. (1998). Extractive fermentation for enhanced propionic acid production from lactose by Propionibacterium acidipropionici. Biotechnology Progress, 14(3), 457–465.
Len, A. C., Harty, D. W., & Jacques, N. A. (2004). Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology, 150(5), 1353–1366.
Liu, L., Zhu, Y., Li, J., Wang, M., Lee, P., Du, G., et al. (2012). Microbial production of propionic acid from propionibacteria: Current state, challenges and perspectives. Critical Review in Biotechnology, 32(4), 374–381.
Lu, P., Ma, D., Chen, Y., Guo, Y., Chen, G. Q., Deng, H., et al. (2013). l-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Research, 23(5), 635–644.
Martínez-Campos, R., & de la Torre, M. (2002). Production of propionate by fed-batch fermentation of Propionibacterium acidipropionici using mixed feed of lactate and glucose. Biotechnology Letters, 24(6), 427–431.
Mashego, M., Wu, L., Van Dam, J., Ras, C., Vinke, J., Van Winden, W., et al. (2004). MIRACLE: Mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnology and Bioengineering, 85(6), 620–628.
Owen, O. E., Kalhan, S. C., & Hanson, R. W. (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. Journal of Biological Chemistry, 277, 30409–30412.
Parizzi, L. P., Grassi, M. C. B., Llerena, L. A., Carazzolle, M. F., Queiroz, V. L., Lunardi, I., et al. (2012). The genome sequence of Propionibacterium acidipropionici provides insights into its biotechnological and industrial potential. BMC Genomics, 13(1), 562.
Piveteau, P. (1999). Metabolism of lactate and sugars by dairy propionibacteria: A review. Le Lait, 79(1), 23–41.
Pluskal, T., Castillo, S., Villar-Briones, A., & Orešič, M. (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics, 11(1), 395–405.
Ruhal, R., & Choudhury, B. (2012). Use of an osmotically sensitive mutant of Propionibacterium freudenreichii subspp. shermanii for the simultaneous productions of organic acids and trehalose from biodiesel waste based crude glycerol. Bioresource technology, 109, 131–139.
Scalbert, A., Brennan, L., Fiehn, O., Hankemeier, T., Kristal, B. S., van Ommen, B., et al. (2009). Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics, 5(4), 435–458.
Senouci-Rezkallah, K., Schmitt, P., & Jobin, M. P. (2011). Amino acids improve acid tolerance and internal pH maintenance in Bacillus cereus ATCC14579 strain. Food Microbiology, 28(3), 364–372.
Suwannakham, S., Huang, Y., & Yang, S. T. (2006). Construction and characterization of ack knock-out mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnology and Bioengineering, 94(2), 383–395.
Suwannakham, S., & Yang, S. T. (2005). Enhanced propionic acid fermentation by Propionibacterium acidipropionici mutant obtained by adaptation in a fibrous-bed bioreactor. Biotechnology and Bioengineering, 91(3), 325–337.
Taymaz-Nikerel, H., De Mey, M., Ras, C., ten Pierick, A., Seifar, R. M., Van Dam, J. C., et al. (2009). Development and application of a differential method for reliable metabolome analysis in Escherichia coli. Analytical Biochemistry, 386(1), 9–19.
Thierry, A., Deutsch, S. M., Falentin, H., Dalmasso, M., Cousin, F. J., & Jan, G. (2011). New insights into physiology and metabolism of Propionibacterium freudenreichii. International Journal of Food Microbiology, 149(1), 19–27.
Want, E. J., Nordström, A., Morita, H., & Siuzdak, G. (2007). From exogenous to endogenous: The inevitable imprint of mass spectrometry in metabolomics. Journal of Proteome Research, 6(2), 459–468.
Wood, H. (1981). Metabolic cycles in the fermentation by propionic acid bacteria. Current Topics in Cellular Regulation, 18, 255–287.
Wu, G. (2009). Amino acids: Metabolism, functions, and nutrition. Amino Acids, 37(1), 1–17.
Xia, M., Huang, D., Li, S., Wen, J., Jia, X., & Chen, Y. (2013). Enhanced FK506 production in Streptomyces tsukubaensis by rational feeding strategies based on comparative metabolic profiling analysis. Biotechnology and Bioengineering, 110, 2717–2730.
Yin, X., Madzak, C., Du, G., Zhou, J., & Chen, J. (2012). Enhanced alpha-ketoglutaric acid production in Yarrowia lipolytica WSH-Z06 by regulation of the pyruvate carboxylation pathway. Applied Microbiology and Biotechnology, 96(6), 1527–1537.
Zeppa, G., Conterno, L., & Gerbi, V. (2001). Determination of organic acids, sugars, diacetyl, and acetoin in cheese by high-performance liquid chromatography. Journal of Agricultural and Food Chemistry, 49(6), 2722–2726.
Zhang, A., & Yang, S. T. (2009a). Engineering Propionibacterium acidipropionici for enhanced propionic acid tolerance and fermentation. Biotechnology and Bioengineering, 104(4), 766–773.
Zhang, A., & Yang, S. T. (2009b). Propionic acid production from glycerol by metabolically engineered Propionibacterium acidipropionici. Process Biochemistry, 44(12), 1346–1351.
Zhu, Y., Li, J., Tan, M., Liu, L., Jiang, L., Sun, J., et al. (2010). Optimization and scale-up of propionic acid production by propionic acid-tolerant Propionibacterium acidipropionici with glycerol as the carbon source. Bioresource Technology, 101(22), 8902–8906.
Zhuge, X., Liu, L., Shin, H., Chen, R. R., Li, J., Du, G., et al. (2013). Development of a Propionibacterium-Escherichia coli shuttle vector as a useful tool for metabolic engineering of Propionibacterium jensenii, an efficient producer of propionic acid. Applied and Environmental Microbiology, 80, 4595–4602.
Acknowledgments
This work was financially supported by the 973 Program (2013CB733902), 111 Project (111-2-06), 863 Program (2011AA100905), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflict of interest
The authors declare that they have no conflict of interests.
Compliance with ethical requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guan, N., Li, J., Shin, Hd. et al. Comparative metabolomics analysis of the key metabolic nodes in propionic acid synthesis in Propionibacterium acidipropionici . Metabolomics 11, 1106–1116 (2015). https://doi.org/10.1007/s11306-014-0766-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-014-0766-3