Abstract
Hybridization is common for many forest trees, where weak barriers to reproduction obscure species boundaries. We characterized the genomic structure of Picea populations comprising three species spanning two well-known contact zones, the Picea sitchensis × Picea glauca and the P. engelmannii × P. glauca hybrid zones, using a set of 71 candidate-gene single nucleotide polymorphisms. The genetic structure of populations suggests a complex genomic architecture shaped by interspecific gene flow and strong environmental selection, with increased genetic diversity in hybrids. The presence of admixture among all three species suggests that three-way hybrids with mixed ancestry occur where species ranges overlap in transitional environments. Significant clinal variation and associations with climatic variables (including continentality, temperature, and precipitation) differ between hybrid zones, indicating that individual species and their hybrids are adapted to distinct environmental niches. Allele–environmental association analysis revealed that most of the candidate genes with evidence of selection were unique to either the Sitka × white or the Engelmann × white hybrid zones, with few shared between these zones. Management of these widespread and diverse gene pools will be best served through development of climate-based seed transfer, with recommended seed sources informed by a combination of genetic and climatic information for future climates.
Similar content being viewed by others
Introduction
Populations adapt to changing conditions through selection on standing genetic variation or on new mutations (Barrett and Schluter 2008). Where the accumulation of new mutations is limited, however, standing genetic variation provides an important source of variation through which populations may adapt to changing selective pressures (Hellmann and Pineda-Krch 2007). Within long-lived forest trees, which exhibit markedly low mutation rates (Neale and Savolainen 2004), standing genetic variation is critical for adaptation to changing conditions (Petit and Hampe 2006). Indeed, spatially and temporally varying selection pressures associated with long-lived, widely distributed forest tree species point towards the importance of standing genetic variation in providing the necessary variation to adapt to changing conditions (Lexer et al. 2004). Interspecific hybridization and introgression result in increased genetic variation, providing an additional mechanism through which standing genetic variation may be maintained across populations, facilitating rapid adaptation to new environments (Aitken et al. 2008; Alberto et al. 2013; Hoffmann and Sgro 2011; Martinsen et al. 2001).
Hybridization is a common phenomenon for many plant taxa, including forest trees. When hybridizing species occur within the same geographical area, weak barriers to reproduction may obscure species boundaries, presenting challenges for individual species management and necessitating the management of species complexes (Jones et al. 2013). Despite a high propensity for introgression, many forest tree species that interbreed maintain morphological, physiological, genetic, and ecological distinctions (Mir et al. 2006; Whittemore and Schaal 1991). A classic example, oaks, represent some of the best studied forest tree hybrid zones, where species integrity is maintained through a combination of genetic and environmental selection despite rampant interspecific gene flow (Lepais and Gerber 2010; Moran et al. 2012; Whittemore and Schaal 1991). These indistinct barriers to reproduction suggest that species-specific mate preference may hold less importance for many long-lived tree species than other plants, or may have had fewer generations to evolve in allopatry or sympatry (Petit et al. 2013).
The extent to which single nucleotide polymorphisms (SNPs) involved in adaptation to climate extends across populations, geographic ranges, and species has great bearing on the transferability of adaptive markers. This remains a key challenge as investigators begin to transfer knowledge from large studies using model systems to natural populations, environments, and non-model species (Grattapaglia et al. 2011; Pavy et al. 2013). Relationships between SNPs and locally adaptive phenotypes may break down: (1) if SNPs are linked to causal polymorphisms rather than causal to adaptive variation; (2) if loci or traits under selection for local adaptation differ with geographic location, i.e., conditional neutrality, with polymorphisms adaptive in some environments or genetic backgrounds but neutral in others; or (3) if SNPs are polymorphic in some populations or species and fixed in another. For example, genome-wide association studies of diseases in humans have found that associations between phenotypes and markers are often in the same direction but differ in strength among human populations, likely due to the associations between marker SNPs and causal variants (Carlson et al. 2013). In natural populations of Arabidopsis thaliana, some SNPs are selectively neutral in certain geographic regions but involved in local adaptation in others, suggesting a nuanced relationship between environment and adaptive genetic variation (Anderson et al. 2013; Fournier-Level et al. 2011). Identification of both shared and non-shared SNPs that may be adaptive across populations, environments, and species will inform predictions concerning the evolutionary trajectory of plant populations, with applications that extend to agricultural, forestry, genetic conservation, and climate change (Bergelson and Roux 2010).
In this study, we characterize the genetic structure of hybrid populations involving three Picea species, P. glauca (Moench) Voss (white spruce), P. sitchensis (Bong.) Carr (Sitka spruce), and P. engelmannii Parry ex Engelm. (Engelmann spruce). Each of these species is adapted to a distinct environmental niche in western North America. Sitka spruce thrives in temperate rainforests within the maritime climate of the Pacific coast. White spruce occupies lower elevation sub-boreal and boreal forests of the interior of British Columbia. Engelmann spruce is found in the interior region in high-elevation montane and subalpine forests (Ketcheson et al. 1991; Pojar et al. 1991). These species are involved in two well-known contact zones in British Columbia that both have white spruce as a parental species: the Sitka × white spruce (SxW) hybrid zone and the Engelmann × white spruce (ExW) hybrid zone. Our previous work has investigated genetic structure within each of these two-species hybrid zones separately (De la Torre et al. 2014a,b,c; Hamilton and Aitken 2013; Hamilton et al. 2013a,b), but the extent of three-way hybridization has never been assessed. A recent study by Pavy et al. (2013) found that 64 % of the 15,388 polymorphic SNPs identified in white spruce from eastern Canada shared polymorphisms with a small sample of Engelmann–white spruce hybrids (ExW) in western Canada, but that only 22 % of these white spruce polymorphisms were shared in a small sample of Sitka spruce. While these species may share polymorphisms, the extent to which those genetic variants are important to local adaptation within one or more species or hybrid complex remains unknown.
Given the different climatic niches of the three species, this research aims to evaluate introgression that may contribute to local adaptation across transitional environments within the species complex. The objective of this research is to (1) determine the presence and extent of introgression among the three spruce species, (2) compare broad-scale genetic clines with spatial and climatic variables to determine the extent to which species integrity is maintained through different environmental selection pressures across different parts of the species complex, and (3) compare fine-scale SNP-climate statistical associations to determine whether the same single nucleotide polymorphisms in candidate genes are associated with climate-related adaptation within the SxW and ExW hybrid zones. We interpret the relevance of these results for climate-based forest management and conservation.
Materials and methods
Sampling
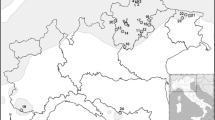
Newly flushed needle tissue was sampled in 2009 from five common garden experiments and clonal archives previously established by the British Columbia (BC) Ministry of Forests, Lands and Natural Resources Operations. Common gardens with different experimental designs were sampled to evaluate genotypes from the Sitka–white spruce (SxW) zone of introgression along the Nass and Skeena rivers of northwestern BC (described in Hamilton et al. (2013b)) and the Engelmann–white spruce (ExW) zone of introgression in the southern interior of BC (described in De la Torre et al. (2014a)). For the SxW zone, six progenies from each of three to seven maternal trees from 29 locations across the introgression zone were sampled, along with allopatric populations representing pure Sitka spruce (HG) and pure white spruce (ENA), for a total of 721 trees (Table 1, Fig. 1). For the ExW hybrid zone, four progenies from 50 seed parents from each of five regional seed planning zones (WK, EK, MR, QL, and PG) were sampled, along with allopatric populations representing pure white spruce (FN), and Engelmann spruce (E1, E2, and E3), for a total of 771 trees (Table 1, Fig. 1). While the sampling strategy varied geographically based on common garden design and genetic materials available, the geographic and environmental representation is broad, spanning both hybrid zones while including pure parental reference populations, in total comprising 1492 genotypes originating from geo-referenced locations (Fig. 1).
Map of origins of individuals (red circles) sampled across the introgression zones among Sitka spruce, white spruce, and Engelmann spruce in British Columbia. Shading indicates the geographic range of Picea glauca (dark gray), P. sitchensis (medium gray), and P. engelmannii (light gray) and overlapping regions within the Pacific Northwest
Genotyping
Newly flushed lateral shoots were collected between June and July 2009, flash-frozen in the field, and transported in liquid nitrogen for subsequent genetic analysis. DNA was extracted using a modified cetyl trimethylammonium bromide protocol (Doyle and Doyle 1990) from 50 mg of tissue ground to a fine powder using Mixer Mill MM 400 (Retsch). To select candidate genes for use across the three spruce species, pure Sitka (N = 1088), white (N = 40), and Engelmann (N = 40) spruce were genotyped for 1536 SNPs identified from candidate genes involved in timing of bud set or fall cold-hardiness in Sitka spruce (Holliday et al. 2008), resistance to the white pine shoot tip weevil (Pissodes strobi; Porth et al. 2011), and growth in white spruce (Pavy et al. 2008; Pelgas et al. 2011). Of these, 71 SNPs that exhibited cross-species amplification were selected for genotyping across the two major zones of introgression, SxW and ExW. Selected SNPs were genotyped using the Illumina bead array platform in conjunction with the GoldenGate allele-specific assay (Fan et al. 2006; Shen et al. 2005). The same genetic materials, but different subsets of SNPs were used in Hamilton et al. (2013a,b) and in De la Torre et al. 2014b. These 71 SNPs were informative across both hybrid zones and met all quality standards based on Hamilton et al. (2013b). Annotation and location of these SNPs in the genome of white spruce can be found in Table S4. SNPs were genotyped in a total of 1492 individuals across 40 populations.
Genomic structure analyses
Preliminary assessment of the genetic relationships among the three pure parent species, along with estimates of pairwise genetic distances between admixed and pure populations, was performed in GenAlEx using data for 71 SNPs and 1492 individuals (Peakall and Smouse 2006). Heterozygosity was calculated for all populations and species, along with estimates of genetic differentiation (F ST) between species. While we recognize that maternal family structure implicit in the design of our experiment may impact allele frequency estimates, we assume that mixing families with a large number of open-pollinated seedlings may yield populations in Hardy–Weinberg equilibrium (Streiff et al. 1999). Based on previous research that found large Sitka spruce populations in the region of contact with on average18–20 effective pollen parents per mother tree (Mimura and Aitken 2007), we expect open-pollinated progeny in the populations we sampled to capture a large sample of paternal haplotypes with minimal bias associated with relatedness.
We used the Bayesian clustering approach implemented in Structure version 2 (Pritchard et al. 2000) to identify genetic clusters and estimate ancestry based on assignment to genetic clusters using the same 71 SNPs and 1492 individuals. Structure estimates the posterior probability (q) of assignment to K genotypic clusters. Using multilocus genotypes based on the assignment to K genetic clusters, we estimated the proportion of each individual’s genome that was derived from a given genetic population (K; Falush et al. 2003). Structure runs were carried out using the admixture model assuming correlated allele frequencies, with a burn-in step of 50,000, followed by 100,000 Markov chain Monte Carlo iterations. Ten replicate runs of models using genotypic clusters (K) from 1 to 10 were used. We evaluated the relative support for each value of K based on the mean ln P[D] and ΔK (Evanno et al. 2005).
To complement this analysis, we conducted a centered principal components analysis (PCA) and discriminant analysis of principal components (DAPC) for the same 71 SNPs using the “adegenet” package v. 1.3-1 (Jombart 2008) in R v.3.0.3 (R Development Core Team 2014). While both Structure and PCA/DAPC are suitable for analyzing population genetic structure, the multivariate analysis does not make population genetic model assumptions of Hardy–Weinberg equilibrium and no linkage disequilibrium, and may be more suitable where complex hierarchical population structure is observed (Kanno et al. 2011). PCA provides a useful validation of Bayesian clustering, particularly where population sampling may be limited or the number of genetic clusters identified using Bayesian approaches does not correspond to biologically meaningful populations (Fogelqvist et al. 2010; Francois and Durand 2010). DAPC combines the benefits of principal components with those of discriminant analysis, ensuring that variables are uncorrelated while maximizing variation between groups and minimizing within-group variation (Jombart et al. 2010). For both of these analyses, we included an a priori identification of population groups for visualization: Typic Sitka, white, and Engelmann spruce populations comprised three distinct groups, while all other populations were identified as either SxW or ExW. The DAPC approach uses a discriminant analysis of transformed data from the principal components analysis. We retained 60 principal components for the DAPC and analyzed all four resulting discriminant functions. Following this, we identified those loci that had the highest contributions to DAPC1 and DAPC2 loadings.
Genetic clines between hybrid zones
The geographic coordinates (latitude, longitude, and elevation) of individual trees sampled were input into ClimateWNA (Wang et al. 2012) to estimate values for seasonal and annual climatic variables. Previous research within each hybrid zone allowed us to identify climate variables that exhibited strong associations with genetic ancestry within each hybrid zone, including mean annual temperature, mean annual precipitation, continentality (difference between mean coldest month temperature and mean warmest month temperature), mean coldest month temperature, and precipitation as snow (De la Torre et al. 2014c; Hamilton and Aitken 2013; Hamilton et al. 2013b). We assessed the relationships between the first two genetic principal components (PC1 and PC2) that summarize the genetic relatedness among the three spruce species for the 71 SNPs and individual climate variables, excluding the Eastern North American white spruce population due to its geographic remoteness. In addition, we tested genetic relationships with elevation, as elevation reflects many co-varying environmental factors, and has previously been shown to differentiate between white and Engelmann spruce habitat (De la Torre et al. 2014b).
Genetic clines were estimated for PC1 and PC2 using nonlinear regressions (Arnold 1997) implemented in R 3.0.3 (R Development Core Team 2014), excluding the distant ENA population based on methods described in Hamilton et al. (2013b). Genetic cline parameters were estimated for the SxW and ExW zones separately for each climatic variable because of the differences in climatic niche space between these zones.
Environmental association analysis
We used LFMM to identify loci that may be targets of environmental selection within each hybrid zone. Population history and demography may influence neutral population structure and can confound signatures of natural selection from loci that exhibit strong associations with environmental variables (Eckert et al. 2010). Latent Factor Mixed Models (LFMM, version 1.1) uses hierarchical Bayesian mixed models to detect correlations between environmental and genetic variation while inferring background levels of population structure, leading to a decrease in the number of false-positive associations. LFMM does not require a priori identification of selectively neutral loci before testing for associations with environmental factors (Frichot et al. 2013). Separate analyses were performed for the ExW and SxW zones to allow for comparison of environment–SNP associations between the zones. All candidate gene SNPs were tested for associations with the same five climatic variables used in the genetic cline analysis (above).
SNPs highly ranked in LFMM lists with high z-scores and significant p values (p < 10−4) were strong candidates for selection. Bonferroni correction for multiple testing was applied to all comparisons. Significant SNP (p < 0.0001) associations with each environmental variable were compared between the two hybrid zones to determine if the same SNPs and associated climatic variables were important to adaptation within both hybrid zones. LFMM runs were carried out using 50,000 iterations, tested for different latent factor values (K = 2–7), and repeated several times to ensure model convergence.
Results
Genomic structure of a spruce contact zone
Genetic relationships among the three parent species, estimated using 71 SNP markers with GenAlEx (Peakall and Smouse 2006) indicate greater genetic divergence between Sitka and Engelmann spruce (F ST = 0.419), and between Sitka and white spruce (F ST = 0.476), than between the more closely related white and Engelmann spruce (F ST = 0.200), supporting the current spruce phylogeny (Ran et al. 2006). Populations identified a priori as SxW or ExW had higher heterozygosity (H e; averaging 0.321 and 0.288, respectively) than the typic population samples (Sitka spruce, 0.234; white spruce, 0.170; and Engelmann spruce, 0.194).
Using the Bayesian approach in Structure to identify genetic populations, we observed strong support for the presence of two genetic clusters for both ln P[D] and ΔK (Fig. S1; (Earl and von Holdt 2012; Evanno et al. 2005). These two clusters appear to distinguish Sitka spruce from the genetic cluster containing both Engelmann and white spruce (K = 2; Fig. 2a). Given that K = 3 is more biologically representative of the sampled populations, as evidenced by our other analyses (below), we compared the Bayesian genetic structure of all populations at both K = 2 and K = 3 (Fig. 2a and b, respectively). With three genetic clusters, Sitka spruce, white spruce, and Engelmann spruce are all clearly distinguished, and their hybrids show various combinations of ancestry among these species (Fig. 2b). Trees with ancestry from more than one species at K = 3 are also identified in the modified triplot output from Structure (Fig. 3.). Individuals representing the three parental species are plotted at the triangle apices, those individuals that share ancestry between two species according to Structure are plotted along the sides of the triplot between those species, and individuals with ancestry from all three species are plotted towards the interior.
Genetic structure of Sitka, white, Engelmann, and admixed spruce populations estimated using Structure with 71 SNP loci for K = 2 (a) and K = 3 genetic clusters (b). Each vertical bar represents an individual, and populations are separated by a vertical black line and labeled according to population codes in Table 1. At K = 2, we interpret green and red bars as differentiating between Sitka spruce and combined Engelmann/white spruce ancestry, respectively. At K = 3, we interpret the red as Engelmann and the blue as indicative of white spruce ancestry
Triangular plot for ancestry assignment based on Structure where K = 3 for individuals representing three pure parent species: Sitka spruce, white spruce, and Engelmann spruce. Triangle vertices represent an assignment probability of zero to one to two of the three species: Sitka–white (SxW) (left), Engelmann–white (ExW) (right), and Sitka–Engelmann (lower) spruce. Individuals are identified based on a priori classification to population groups as either pure Sitka spruce (black), pure Engelmann spruce (blue), pure white spruce (red), admixed Sitka–white spruce (SxW) individuals (yellow), admixed ExW individuals (green)
Most admixed individuals shared ancestry from two species, either Sitka and white spruce (SxW), or Engelmann and white spruce (ExW). However, a small number of individuals had varying levels of introgression from all species, suggesting the presence of three-species hybrids. Sixty-nine individuals from the SxW hybrid zone had 10–20 % ancestry from Engelmann spruce, while 16 had 20–30 % and 16 individuals had 30–60 % Engelmann ancestry, from a total of 721 individuals sampled in the SxW hybrid zone. These three-species hybrids are plotted towards the center of the triplot (Fig. 3). Populations from the SxW hybrid zone that are geographically proximal to the ExW hybrid zone (Cedarvale-Low, Kiteen-Low, Kitwanga Lake-Mid, Moricetown, Skeena Crossing, and Talchako River-High) had on average between 10 and 17 % Engelmann ancestry and average H e of 0.39 (Table 1, Table S1). Some degree of introgression from Sitka spruce into those geographically proximal ExW populations has also been found; with 47 individuals having 10–20 % Sitka spruce ancestry and 4 individuals having 20–30 % Sitka ancestry out of a total of 771 individuals sampled in the ExW hybrid zone. The Prince George population had an average of 7 % Sitka spruce ancestry, the highest found in the ExW hybrid zone.
PCA and DAPC were used to further explore the genetic distribution of spruce ancestry within this complex, reducing results from the 71 SNPs to a few canonical vectors. The majority of variance in the PCA was represented by the first principal component, which explains 52.63 % of the total variance, and separates Sitka spruce from the white/Engelmann spruce complex. The second principal component explains just 5.55 % of variation, but clearly separates white and Engelmann spruce (Fig. 4a). These results are elaborated on using DAPC, which retains a number of principal components for discriminant analysis. Unlike PCA where each eigenvalue indicates the proportion of variance explained per PC, the DAPC eigenvalues correspond to a ratio of between to within group variance calculated for each discriminant function. Consistent with the PCA, the greatest ratio of between to within group variation is observed in the first eigenvalue (5221.66), pointing towards differentiation between Sitka spruce and the Engelmann/white spruce complex (Fig. 4b). The second eigenvalue (359.62) differentiates Engelmann and white spruce. In both cases, those individuals sampled from either the SxW or ExW zone of introgression appear intermediate to the pure parental species. While SxW individuals appear more Sitka spruce-like in both the PCA and DAPC analysis, ExW individuals appear more Engelmann spruce-like within the PCA, but more white spruce-like within the DAPC.
Genetic composition of 1492 individuals representing five classes of species and hybrids based on multivariate analyses of 71 SNPs: Sitka, white, and Engelmann spruce, Sitka–white hybrids (SxW), and Engelmann–white hybrid (ExW) arranged along axes 1 and 2 of (a) PC1 and PC2 scores from a principal components analysis, which explain 52.63 and 5.55 % of the total genetic variation, respectively, distinguishing primarily between Sitka and Engelmann/white spruce and (b) a discriminant analysis of principal components (DAPC), which further differentiates Engelmann from white spruce
Of the ten loci that had the highest contributions to the DAPC analysis (Table S2), five loci contributed disproportionately to DAPC1, and five loci contributed disproportionately to DAPC2 (Fig. S2). Consequently, the genetic structure of the groups is disproportionately influenced by loci that discriminate Sitka spruce primarily from Engelmann and white spruce (DAPC1) and Engelmann spruce from white spruce (DAPC2), respectively. Interestingly, two of the loci (213_330_S and 213_72_S) have previously been identified within the SxW hybrid zone as exhibiting excess genetic differentiation, suggesting that they may be candidates for divergent selection (Hamilton et al. 2013a)
Genetic clines
Genetic clines were estimated for the first and second principal components of the genetic structure analysis with climate and geographic variables within each hybrid zone (Fig. S3, Table 2). As genetic clines associated with environmental gradients were not assumed to extend across both hybrid zones, individual cline parameters were analyzed within and then compared between the SxW and ExW zone (Fig. S3, Table 2).
Significant clines were observed for all climate and distance variables in association with PC1 within the SxW zone (Table 2, Fig. S3A). In contrast, weak or no clinal variation was observed within the ExW zone for the same climate and distance variables with PC1. These results suggest that the first principal component is primarily associated with the environmental gradient that influences the genetic structure of the SxW zone extending from a maritime to continental climate. However, PC2 was associated with several climatic and geographic variables in both the ExW and in the SxW spruce zone (Table 2, Fig. S3B). Associations between PC2 scores and climatic variables in the two hybrid zones were similar for mean annual precipitation, continentality, and precipitation as snow, but were in the opposite direction for mean annual temperature, mean coldest month temperature, and elevation. This suggests that the environmental gradients associated with PCA2, discriminating primarily between Engelmann spruce and white spruce, differ from those distinguishing Sitka from white spruce.
To effectively visualize the complex genetic and climatic patterns across the two hybrid zones, we used a three-dimensional plot to illustrate the relationship between genetic variation for PC1 and PC2 with climatic variation based on continentality and mean coldest month temperature (Fig. 5). Analysis of individual hybrid zones suggested that the primary climatic variables that distinguish Sitka spruce from the ExW complex differ from those that separate white spruce from Engelmann spruce. These results suggest that the genetic variation comprising PC1 and PC2 is structured along different environmental gradients across this Picea complex.
Three-dimensional relationship between PC1 and PC2 scores from a principal component analysis of genotypes for 71 SNP loci for 1492 individuals spanning the contact zone between Sitka, white, and Engelmann spruce and admixed individuals, and climatic variables continentality (°C) and mean coldest month temperature
Environmental association analyses
Associations between allele frequencies and climate variables were estimated for each hybrid zone separately (SxW and ExW) using LFMM. The distribution of z-scores was similar in both hybrid zones, with most (63–100 %) of the SNPs showing z-scores between 0 and 5, some (0–37 %) having z-scores between 5 and 10, and very few (0–1 %) having z-scores over 10 (Table 3). Most of the SNPs with higher z-scores were associated with more than one environmental variable within each of the hybrid zones. The distribution of the z-scores points towards different environmental selection pressures in each of the two hybrid zones. While mean annual precipitation, continentality, and mean coldest month temperature seem to be the most important environmental variables shaping the SxW zone, precipitation as snow and mean annual precipitation have the strongest associations with allele frequencies in the ExW zone (Table 3).
We identified SNPs that exhibited significant associations with climatic variables in each hybrid zone as those with a z-score higher than 5, corresponding to p < 10−5 following a Bonferroni correction for a type I error and α = 0.001. When several SNPs met this criterion, only the first three SNPs with z-score > 5 were considered, following Eckert et al. (2010). The relatively small number of SNPs used in this study may be not enough to correct for all neutral population structure if there is undetected population stratification in the hybrid zones. By taking only the SNPs highly ranked in the LFMM lists, we hope to reduce the number of spurious associations.
We found three candidate gene SNPs that exhibited strong associations with mean annual temperature, six with continentality, six with mean coldest month temperature, six with mean annual precipitation, and six with precipitation as snow (Table 3). Only one SNP within the SxW hybrid zone (133_39_S) exhibited a z-score higher than 10. Three SNP loci (127_273; 198_447 and 45_1067) exhibited z-scores > 5 within both hybrid zones (Table S3); however, there were no SNPs identified as potential targets of selection in both the SxW zone and the ExW zone.
Discussion
The genetic structure of populations spanning the introgression zones of Sitka, white, and Engelmann spruce suggests species integrity is likely maintained through environmental selection in parental habitats, but hybridization may facilitate fine-scale adaptation along climatic gradients. Significant clinal variation along environmental gradients suggests that the genetic structure of both pure species and admixed individuals may be strongly influenced by climate. However, the climatic factors influencing population structure differ between the two dominant hybrid zones, SxW and ExW. The SNPs that may contribute to local adaptation and are associated with those climatic factors differ between zones, pointing towards an independent genetic basis for adaptation across different environments. These ecological transition zones between species with weak barriers to reproduction have resulted in populations with higher genetic variation than parental populations, which may contribute to adaptation across diverse environments.
Genomic structure of a spruce species complex
Barriers to reproduction among these three species appear to be weak. While introgression is mostly observed within the ExW and SxW zones, we detected evidence of variable levels of introgression from Engelmann spruce into several geographically proximal SxW admixed populations (Fig. 2). Varied amounts of Engelmann spruce ancestry (10–60 %) were mainly observed in Cedarvale-Low, Kiteen-Low, Kitwanga Lake-Mid, Moricetown, Skeena Crossing, and Talchako River-High populations. In addition, small amounts of Sitka spruce ancestry (7 %) were observed in PG, an ExW hybrid population of central British Columbia that is geographically proximal to the SxW hybrid zone. While SxW and ExW hybrids are common and dominate their respective zones, our estimates of admixture suggest a small number of individuals genotyped (n = 16) shared 30–60 % ancestry from both Sitka spruce and Engelmann spruce (Fig. 2). However, these results do not necessarily suggest that there is selection against introgression between Sitka and Engelmann spruce. This small number of apparent Sitka–Engelmann hybrids may have resulted from gaps in the geographic sampling in the contact area between the two hybrid zones, geographic isolation between pure parental species, or variation in timing of reproductive phenology.
The detection of some individuals with shared ancestry from all three species, with at least 20 % ancestry from each of Sitka, white, and Engelmann spruce, is consistent with previous morphological and cytoplasmic studies (Sutton et al. 1994; Richardson et al. 2000) revealing a complex relationship among these spruce species. Our previous research within the two dominant hybrid zones SxW and ExW provided strong evidence for asymmetrical gene flow in both zones, with an excess of Sitka spruce ancestry in the SxW zone, and an excess of Engelmann spruce ancestry within the ExW zone, relative to white spruce (De la Torre et al. 2014b; Hamilton et al. 2013b). White spruce may provide a conduit for gene flow between Engelmann spruce and Sitka spruce. Despite being less abundant than the other spruce species in both zones, this gene flow likely provides an important source of genetic variation within ecotonal environments. This may have important consequences with respect to adaptation if asymmetrical gene flow within each hybrid zone limits adaptation or results in the loss of locally adapted white spruce alleles (Alberto et al. 2013).
While we observed few individuals with ancestry from only Sitka and Engelmann spruce, and others with ancestry from all three species, shared polymorphisms may influence our estimates of the genetic contributions for each species. Several studies have shown extensive allele sharing for nuclear and organelle genes among Engelmann, white, and Sitka spruce, although the degree of sharing differs between pairs of species (Bouille and Bousquet 2005; Bouillé et al. 2011; Pavy et al. 2013). Phylogenies group Engelmann spruce and white spruce as sister species, with Sitka spruce being more distantly related (Ran et al. 2006). This is concordant with our analysis using Structure, which provides strong support for two major genetic clusters across the three species, separating Sitka spruce from white and Engelmann spruce (Fig. 2 and S1). Ran et al. (2006) examined chloroplast phylogenies in North American spruce and found that Engelmann and white spruce formed a sister clade at the terminus of chloroplast phylogenies, whereas Sitka spruce formed a more basal node within the topology (Ran et al. 2006).
Although estimates of genetic differentiation based on SNPs are, on average, higher than those based on microsatellites (De la Torre et al. 2014a; Hamilton and Aitken 2013), overall patterns of differentiation between species are similar among marker types. Our results supporting K = 2 genetic populations for these 71 SNPs mainly reflects the greater genetic divergence between Sitka and Engelmann (F ST = 0.419), and Sitka and white (F ST = 0.476) than between the sister species white and Engelmann spruce (F ST = 0.2). Assignment tests such as Structure may underestimate the number of genetic clusters where there is hierarchical structure, large variances in sample size, or a high degree of allele sharing between species (Fogelqvist et al. 2010). Results from the PCA and DAPC analyses suggest that K = 3, separating Sitka, white, and Engelmann spruce, better represents the biology within this species complex as white and Engelmann spruce are genetically distinct species (De la Torre et al. 2014b; Haselhorst and Buerkle 2013; Nkongolo et al. 2005).
The PCA and DAPC provide additional information about population genetic structure within this complex. The majority of variation is explained by the differentiation of Sitka spruce from the Engelmann–white spruce complex (PC1 and DAPC1), while the second axis of both analyses distinguishes Engelmann and white spruce, supporting K = 3 (Fig. 4, Table 2). Two of the five loci that contributed disproportionately to DAPC1, discriminating Sitka spruce from both Engelmann and white spruce, were also identified as F ST outliers in a previous study in the SxW hybrid zone (Hamilton et al. 2013a). These loci, homologous to sphingolipid desaturases, were previously associated with functional roles in adaptation to freezing and dehydration (Chen et al. 2012; Steponkus 1984). Similarly, two of the five loci that contributed disproportionately to DAPC2 have been identified as potentially adaptive loci in recent studies within the ExW hybrid zone (De la Torre et al. 2014a,b; Fig. S2). Both loci have functions associated with adaptation to cold and development of cold hardiness (Dauwe et al. 2012; Holliday et al. 2008). As such, genetic variation summarized in DAPC1 and DAPC2 may be disproportionately influenced by those loci that contribute to adaptive differentiation within either the SxW or ExW zone.
Environmental associations
The species included in our analyses have likely been in contact through recurrent episodes of secondary contact, a consequence of range expansions and contractions during the Pleistocene, most recently during (ExW) and following (ExW and SxW) the last glacial maxima (De la Torre et al. 2014b; Hamilton and Aitken 2013). Despite rampant interspecific gene flow, these three species remain differentiated ecologically, morphologically, and genetically. Interspecific gene flow results in populations comprising individuals with mixed ancestry in environments where they are favored, often intermediate to those inhabited by pure parental species. This contributes to locally adapted, stable hybrid swarms, typical of a bounded hybrid superiority model of hybrid zone maintenance (Arnold 1997). The three-dimensional depiction of the hybrid zones (Fig. 5) suggests that while all SNPs are polymorphic in both SxW and ExW zones, the genetic variation comprising PC1 and PC2 is structured along different environmental gradients in the two zones, providing support for different selection pressures acting in each of these zones.
The comparison of genetic clines between these two hybrid zones suggest that while both zones are shaped to some extent by mean annual temperature, the SxW zone is strongly influenced by precipitation, continentality, and mean coldest month temperature, while the ExW hybrid zone is structured along gradients of precipitation as snow and elevation (Table 2). These results suggest Sitka, Engelmann, and white spruce as well as admixed individuals are adapted to distinct climates, although we cannot rule out adaptation to other non-climatic environmental factors, such as soils, photoperiod, light intensity, or the presence of other species (Aitken and Whitlock 2013). Precipitation appears to be a key factor influencing the genetic structure of individuals across the SxW hybrid zone. Sitka spruce is adapted to wet maritime climates, with precipitation values typically exceeding 1000 mm annually, whereas SxW hybrids occupy a narrow transition in precipitation just below this threshold, below which white spruce genotypes predominate (Bennuah et al. 2004; Hamilton et al. 2013b). In contrast, within the continental climate of interior British Columbia, elevation and climatic variables strongly correlated with elevation provide the steep gradient distinguishing Engelmann and white spruce. Engelmann spruce is adapted to persistent snowpacks and short growing seasons within subalpine environments, whereas white spruce is found at lower elevations that are warmer and drier in summer, but can be colder in winter (De la Torre et al. 2014c). These results complement recent observations in Arabidopsis, which found that alleles associated with higher fitness tend to be local alleles associated with particular climatic factors (Fournier-Level et al. 2011; Hamilton et al. 2014), although the distribution of alleles in predominantly outcrossing spruces is less restricted geographically than in inbreeding Arabidopsis.
Fine-scale environmental association analyses suggested that most of the SNPs associated with environmental variables are unique to either the SxW or ExW zone. Some SNPs exhibited associations with several environmental variables within a zone, likely reflecting correlations among environmental variables, but only three SNPs were shared between the zones (Table S3). These results may indicate conditional neutrality of SNP polymorphisms, with different genes or different polymorphisms within genes being targets of selection in different environments or in different genetic backgrounds (Anderson et al. 2013; Hamilton et al. 2013a; Hancock et al. 2011).
A subset of the SNPs detected here were associated with the climate-relevant phenotypic traits bud set timing and cold hardiness in a previous study of Sitka spruce (Holliday et al. 2008) and recent analyses using F ST-outlier analysis and genomic clines in the ExW hybrid zone (De la Torre et al. 2014b; Hamilton et al. 2013a). In addition, a number of the SNPs identified in this study have previously been identified as candidates for selection the SxW hybrid zone. Some of these SNPs exhibited evidence of positive selection in one species genomic background and negative selection in the other (Hamilton et al. 2013a).
Implications for forest breeding and management
These three spruce species span a large range of climatic and ecological conditions, from temperate rainforests to boreal and subalpine environments. Weak barriers to reproduction have allowed for the evolution of heavily introgressed populations adapted to local climates to persist in intermediate habitats. This supports the important role of standing genetic variation in forest trees, where standing genetic variation is implicated over de novo variation in fueling local adaptation to climate in these long-lived species (Aitken et al. 2008; Alberto et al. 2013). Greater genetic diversity in admixed populations should provide these populations with considerable capacity to adapt rapidly to changing conditions. An earlier study predicted that climate change would likely favor white spruce ancestry in the current ExW zone as temperatures become milder at higher elevations (De la Torre et al. 2014c). White spruce ancestry within this hybrid zone is already being increased in several breeding populations as a result of selection for faster tree growth (De la Torre et al. 2014c). With global warming, conditions may become more favorable for Sitka spruce ancestry within the current SxW region, at the expense of white spruce ancestry; however, this will depend on future precipitation regimes (Hamilton et al. 2013b).
Understanding the complex ecological and genetic relationships between introgressant species will improve opportunities for successful management in a changing climate. The management of these widespread and diverse gene pools for future climates will be best served by the development of climate-based seed transfer for reforestation and restoration, with seed source selection informed by a combination of genetic and climatic information (Aitken and Whitlock 2013). Current seed transfer is restricted within geographically rather than climatically delineated seed zones, and selection of seed sources for reforestation in ecological transitional areas remains problematic. Identifying populations to use as seed sources that are optimally adapted to particular environments under new climates should translate into better tree health and productivity, and consequently into greater economic and ecological value.
References
Aitken SN, Whitlock MC (2013) Assisted gene flow to facilitate local adaptation to climate change. Ann Rev Ecol Evol Syst 44:367–388
Aitken SN, Yeaman S, Holliday JA, Wang T, Curtis-McLane S (2008) Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol Appl 1:95–111. doi:10.1111/j.1752-4571.2007.00013.x
Alberto F et al. (2013) Potential for evolutionary responses to climate change—evidence from tree populations. Glob Chang Biol 19:1645–1661
Anderson JT, Lee C, Rushworth CA, Colautti RI, Mitchell-Olds T (2013) Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol 22:699–708
Arnold ML (1997) Natural hybridization and evolution. Oxford University Press, New York
Barrett R, Schluter D (2008) Adaptation from standing genetic variation. Trends Ecol Evol 23:38–44. doi:10.1016/j.tree.2007.09.008
Bennuah SY, Wang T, Aitken SN (2004) Genetic analysis of the Picea sitchensis × glauca introgression zone in British Columbia. For Ecol Manag 197:65–77. doi:10.1016/j.foreco.2004.05.005
Bergelson J, Roux F (2010) Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet 11:867–879
Bouille M, Bousquet J (2005) Trans-species shared polymorphisms at orthologous nuclear gene loci among distant species in the conifer Picea (Pinaceae): implications for the long-term maintenance of genetic diversity in trees. Am J Bot 92:63–73
Bouillé M, Senneville S, Bousquet J (2011) Discordant mtDNA and cpDNA phylogenies indicate geographic speciation and reticulation as driving factors for the diversification of the genus. Picea Tree Genet Genomes 7:469–484. doi:10.1007/s11295-010-0349-z
Carlson CS et al. (2013) Generalization and dilution of association results from European GWAS in populations of non-European Ancestry: the PAGE Study. PLoS Biol 11:e1001661
Chen M, Markham JE, Cahoon EB (2012) Sphingolipid 8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis The. Plant J 69:769–781
Dauwe R, Holliday JA, Aitken S, Mansfield SD (2012) Metabolic dynamics during autumn cold acclimation within and among populations of Sitka spruce (Picea sitchensis). New Phytol 194:192–205
De la Torre AR, Ingvarsson PK, Aitken SN (2014a) Genetic architecture and genomic patterns of gene flow between hybridizing species of Picea. Heredity
De la Torre AR, Roberts DR, Aitken S (2014b) Genome-wide admixture and ecological niche modeling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Mol Ecol 23:2046–2059
De la Torre AR, Wang T, Jaquish BC, Aitken SN (2014c) Adaptation and exogenous selection in a Picea glauca × P. engelmannii hybrid zone: implications for forest management under climate change. New Phytol 201:123–139
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, von Holdt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Eckert AJ, Bower AD, González-Martínez SC, Wegrzyn JL, Coop G, Neale DB (2010) Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae). Mol Ecol 19:3789–3805
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fan J et al (2006) Illumina universal bead arrays. Methods Enzymol 410:57–73. doi:10.1016/s0076-6879(06)10003-8
Fogelqvist J, Niittyvuopio A, Agren J, Savolainen O, Lascoux M (2010) Cryptic population genetic structure: the number of inferred clusters depends on sample size. Mol Ecol Resour 10:314–323
Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM (2011) A map of local adaptation in Arabidopsis thaliana. Science 334:86–89
Francois O, Durand E (2010) Spatially explicit Bayesian clustering models in population genetics. Mol Ecol Resour 10:773–784
Frichot E, Schovile SD, Bouchard G, Francois O (2013) Test for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol 30:1687–1699
Grattapaglia D, Silva-Junior OB, Kirst M, Lima B, Faria DA, Pappas G (2011) High-throughput SNP genotyping in the highly heterozygous genome of Eucalyptus: assay success, polymorphism, and transferability across species. BMC Plant Biol 11:65
Hamilton JA, Aitken SN (2013) Genetic and morphological structure of a spruce hybrid (Picea sitchensis × P. glauca) zone along a climatic gradient. Am J Bot 100:1651–1662
Hamilton JA, Lexer C, Aitken SN (2013a) Differential introgression reveals candidate genes for selection across a spruce (Picea sitchensis × P. glauca) hybrid zone. New Phytol 197:927–938
Hamilton JA, Lexer C, Aitken SN (2013b) Genomic and phenotypic architecture of a spruce hybrid zone (Picea sitchensis × P. glauca). Mol Ecol 22:827–841
Hamilton JA, Okada M, Korves TM, Schmitt J (2014) The role of climate adaptation in colonization success in Arabidopsis thaliana. Mol Ecol
Hancock AM et al (2011) Adaptation to climate across the Arabidopsis thaliana genome. Science 334:83–86
Haselhorst MSH, Buerkle C (2013) Population genetic structure of Picea engelmannii, P. glauca and their previously unrecognized hybrids in the central Rocky Mountains. Tree Genet Genomes 9(3):669–681
Hellmann J, Pineda-Krch M (2007) Constraints and reinforcement on adaptation under climate change: selection of genetically correlated traits. Biol Conserv 137:599–609. doi:10.1016/j.biocon.2007.03.018
Hoffmann AA, Sgro CM (2011) Climate change and evolutionary adaptation. Nature 470:479–485
Holliday JA, Ralph SG, White R, Bohlmann J, Aitken SN (2008) Global monitoring of autumn gene expression within and among phenotypically divergent populations of Sitka spruce (Picea sitchensis). New Phytol 178:103–122. doi:10.1111/j.1469-8137.2007.02346.x
Jombart T (2008) adegenet: a R package for multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jones RC, Steane DA, Lavery M, Vaillancourt R, Potts B (2013) Multiple evolutionary processes drive the patterns of genetic differentiation in a forest tree species complex. Ecol Evol 3:1–17
Kanno Y, Vokoun JC, Letcher BH (2011) Fine-scale population structure and riverscape genetics of brook trout (Salvelinus fontinalis) distributed continuously along headwater channel networks. Mol Ecol 20:3711–3729
Ketcheson MV, Braudmandl TF, Meidinger D, Utzig G, Demarchi DA, Wikeem BM (1991) Interior Cedar-Hemlock Zone. Victoria, BC
Lepais O, Gerber S (2010) Reproductive patterns shape introgression dynamics and species succession within the European white oak species complex. Evolution 65:156–170
Lexer C, Henize B, Alia R, Rieseberg LH (2004) Hybrid zones as a tool for identifying adaptive genetic variation in outbreeding forest trees: lessons from wild annual sunflowers (Helianthus spp). For Ecol Manag 197:49–64. doi:10.1016/j.foreco.2004.05.004
Martinsen GD, Whitham TG, Turek RJ, Keim P (2001) Hybrid populations selectively filter gene introgression between species. Evolution 55:1325–1335
Mimura M, Aitken SN (2007) Increased selfing and decreased effective pollen donor number in peripheral relative to central populations in Picea sitchensis (Pinaceae). Am J Bot 94:991–998
Mir C, Toumi L, Jarne P, Sarda V, Di Giusto F, Lumaret R (2006) Endemic North African Quercus afares Pomel originates from hybridisation between two genetically very distant oak species (Q. suber L. and Q. canariensis Willd.): evidence from nuclear and cytoplasmic markers. Heredity 96:175–184
Moran EV, Willis JH, Clark JS (2012) Genetic evidence for hybridization in red oaks (Quercus sect. Lobatae, Fagaceae). Am J Bot 99:92–100
Neale D, Savolainen O (2004) Association genetics of complex traits in conifers. Trends Plant Sci 9:325–330
Nkongolo K, Michael P, Demers T (2005) Application of ISSR, RAPD, and cytological markers to the certification of Picea mariana, P. glauca, and P. engelmannii trees and their putative hybrids. Genome 48:302–311
Pavy N et al (2008) Enhancing genetic mapping of complex genomes through the design of highly-multiplexed SNP arrays: application to the large and unsequenced genomes of white spruce and black spruce. BMC Genomics 9:21. doi:10.1186/1471-2164-9-21
Pavy N et al (2013) Development of high-density SNP genotyping arrays for white spruce (Picea glauca) and transferability to subtropical and nordic congeners. Mol Ecol Resour 13:324–336
Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pelgas B, Bousquet J, Meirmans PG, Ritland K, Isabel N (2011) QTL mapping in white spruce: gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. BMC Genomics 12:145. doi:10.1186/1471-2164-12-145
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree annual review of ecology. Evol Syst 37:187–214. doi:10.1146/annurev.ecolsys.37.091305.110215
Petit RJ et al (2013) Fagaceae trees as models to integrate ecology, evolution and genomics. New Phytol 197:369–371
Pojar J, Klinka K, Demarchi DA (1991) Coastal Western Hemlock Zone. Victoria, BC
Porth I, Hamberger B, White R, Ritland K (2011) Defense mechanisms against herbivory in Picea: sequence evolution and expression regulation of gene family members in the phenylpropanoid pathway. BMC Genomics 12:608
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ran J-H, Wei X-X, Wang X-Q (2006) Molecular phylogeny and biogeography of Picea (Pinaceae): implications for phylogeographical studies using cytoplasmic haplotypes. Mol Phylogenet Evol 41:405–419. doi:10.1016/j.ympev.2006.05.039
R Development Core Team (2014) R: a language and environment for statistical computer. Vienna. http://www.R-project.org/
Richardson AD, Berlyn GP, Ashton PMS, Thadani R, Cameron IR (2000) Foliar plasticity of hybrid spruce in relation to crown position and stand age. Can J Bot 78:305–317
Shen R et al (2005) High-throughput SNP genotyping on universal bead arrays. Mutat Res Fundam Mol Mech Mutagen 573:70–82. doi:10.1016/j.mrfmmm.2004.07.022
Steponkus PL (1984) Role of the plasma membrane in freezing injury and cold acclimation. Ann Rev Plant Physiol 35: 543–584
Streiff R, Ducousso A, Lexer C, Steinkellner H, Gloessl J, Kremer A (1999) Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Q. petraea (Matt.) Liebl. Mol Ecol 8:831–841
Sutton BCS , Pritchard SC, Gawley JR, Newton CH (1994) Analysis of Sitka spruce - interior spruce introgression in British Columbia using cytoplasmic and nuclear DNA probes. Can J For Res 24:278–285
Wang T, Hamann A, Spittlehouse DL, Murdock TQ (2012) ClimateWNA—high resolution spatial climate data for western North America. J Appl Meteorol Climatol 51:16–29
Whittemore AT, Schaal BA (1991) Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci 88:2540–2544
Acknowledgments
We thank John King and Barry Jaquish from the British Columbia Ministry of Forests, Lands and Natural Resource Operations for establishing the common garden trials and providing material, and Christine Chourmouzis, Lisa Erdle, Nina Lobo, Jon Sweetman, Pia Smets, and Jordan Bemmels for field assistance. We also thank Eric Frichot and Graham Coop for valuable suggestions on the environmental association analyses. Particular thanks to Santiago Gonzalez-Martinez and anonymous referees for thoughtful suggestions that have greatly improved the manuscript. This work was supported by Genome British Columbia, Genome Canada, the Province of British Columbia and the British Columbia Forest Genetics Council (grant to S.N.A.), the Natural Science and Engineering Research Council of Canada (NSERC Discovery grant to S.N.A.), an NSERC Canada Graduate Scholarship to J.H., and UBC Fellowships to J.H and A.R.T.
Data archiving statement
SNP data for the Picea glauca × P. engelmannii hybrid zone is available at Dryad doi:10.5061/dryad.7h65f. Population origin and SNP genotypes for the Picea sitchensis × P. glauca are available at Dryad doi: 10.5061/dryad.s11b6.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S.C. González-Martínez
J. A. Hamilton and A. R. De la Torre contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(XLSX 49 kb)
Table S2
(XLSX 37 kb)
Table S3
(DOCX 130 kb)
Table S4
(XLSX 49 kb)
Fig. S1
Population genetic structure analyses based on ten replicate Structure runs for Sitka, white, Engelmann, and admixed spruce. Mean ln P[D] (dotted line) and K genetic clusters (solid line) for K = 1–10. (PDF 106 kb)
Fig. S2
Loading plot of SNP contributions to the first discriminant principle component function (a) and second discriminant principle component function (b) based on 71 SNPs across the Sitka, white, Engelmann, and admixed populations. The gray line indicates the 95% quantile. (GIF 54 kb)
Fig. S3
(GIF 607 kb)
Rights and permissions
About this article
Cite this article
Hamilton, J.A., De la Torre, A.R. & Aitken, S.N. Fine-scale environmental variation contributes to introgression in a three-species spruce hybrid complex. Tree Genetics & Genomes 11, 817 (2015). https://doi.org/10.1007/s11295-014-0817-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-014-0817-y