Abstract
The evolution of the genetic code in terms of the adoption of new codons has previously been related to the relative thermostability of codon–anticodon interactions such that the most stable interactions have been hypothesised to represent the most ancient coding capacity. This derivation is critically dependent on the accuracy of the experimentally determined stability parameters. A new set of parameters recently determined for B-DNA reveals that the codon–anticodon pairs for the codes in non-plant mitochondria on the one hand and prokaryotic and eukaryotic organisms on the other can be unequivocally divided into two classes – the most stable base steps define a common code specified by the first two bases in a codon while the less stable base steps correlate with divergent usage and the adoption of a 3-letter code. This pattern suggests that the fixation of codons for A, G, P, V, S, T, D/E, R may have preceded the divergence of the non-plant mitochondrial line from other organisms. Other variations in the code correlate with the least stable codon–anticodon pairs.
Similar content being viewed by others
Introduction
The evolution of the genetic code is a classic example of a phenomenon for which there are plethora of hypotheses and a dearth of data. The data and their consistency have been comprehensively reviewed by Trifonov (2000) yet there remains a lack of consensus on the issue. However, any hypothesis must ultimately be consistent with the thermodynamics of the coding process. Central to this process, and more importantly, a quantifiable parameter, is the interaction of the codon with its complementary anticodon. Here I argue that the correlations between codon–anticodon stability and coding variability argue in favour of the evolution of divergent genetic codes accompanied by a transition from a 2-letter triplet code to a 3-letter triplet code.
Codon–Anticodon Thermostability
The concept that the primacy of codon acquisition in the development of the genetic code is dependent on the stability of the codon–anticodon complement was first advanced by Eigen and Schuster (1978). This robust concept was further developed by Trifonov (2000, 2004) who argued that the thermostability rule, when applied to coding triplets, was consistent, with only a few exceptions, with the deduced chronological accretion of codons to the standard code. Strikingly the most ancient amino acids in his derived temporal order mirrored the nine most abundant encoded amino acids in Miller’s recreation of the hypothetical primordial environment (Miller 1953; Miller and Urey 1959). This correlation of thermostability with the chronology of code evolution depends critically on the accuracy of the values assigned to the stability of the individual base steps. Up to recently the favoured experimental method for determining these values for both RNA and DNA was to determine the melting temperature for oligonucleotides of defined length sequence and then to deconvolute the parameters for base steps (i.e., dinucleotides) based on base step composition of the tested sequences (SantaLucia 1998; Xia et al. 1998). However recently a novel enzymatically based method was applied to directly determine the stacking energy of individual base steps in DNA at physiological temperatures (Protozanova et al. 2004). These new values differ significantly from those derived from oligonucleotide studies in that the purine–pyrimidine steps and the pyrimidine–purine steps are relatively more and less stable, respectively. They further show that the principal component of the stability of a base step is the stacking energy. Importantly this set of parameters more accurately predicts both the persistence length of DNA in solution and the untwisting of DNA driven by negative superhelicity (Travers 2006). The question is whether it is legitimate to apply this ordering of base step stability to RNA which exists as a double-helical form entirely in an A-type conformation. However in A-DNA the conformational variation of roll angle follows similar rules to B-DNA i.e., RY low roll, RR/YY, medium positive roll, YR high positive roll (because of purine clash of propeller-twisted base pairs) although average roll angle is more positive and average twist angle lower. Consequently the relative stability of base-step type in A-DNA and thus probably in RNA should in principle follow similar rules to those for B-DNA especially with regard to the less stable YR steps. Another relevant point is that studies on duplex stability are all determined under a particular set of environmental conditions which may, or more probably may not, be the same as those under which the code evolved. Given this caveat, it has to be assumed that the relative stabilities of base steps are maintained over a significant range of conditions.
An important issue when assessing the stability of codon–anticodon interactions is the relative contribution of the two base steps in the three base-pair duplex. In this discussion the first base step, corresponding to XY in an XYZ codon, are considered initially on the grounds that in contemporary biological systems, the third base pair is subject to the ‘wobble’ rules of base pairing (Crick 1966). These allow for more flexibility in pairing – for example, certain purine–purine pairs are allowed – and are accommodated on the ribosome by changes in the conformation of the participating nucleotides (Murphy and Ramakrishnan 2004). Consequently the energetics of any stacking interactions on the preceding base pair would likely differ from experimentally established values using duplex nucleic acids. Additionally, the importance of stacking interactions in stabilising duplex formation argues against the possibility, first advanced by Hartman (1975), that the initial code was singlet in nature.
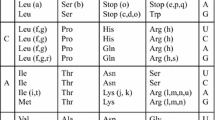
When the base step stability of the first two bases in a codon–anticodon pair is compared to coding capacity several correlations are observed (Table I). In general, as previously noted, the most stable initial base steps correspond the coding boxes containing a single amino acid in the standard code while the less stable ones are associated with boxes containing two or even three outcomes. This correlation is dependent on melting stability and not on stacking stability alone. It should be noted that the difference in melting energies between the base steps GT/AC and GG/CC and also between CT/AG and AT is sufficiently small that the constituents of these pairs, given the discussed uncertainties, are essentially equivalent. Second, again as noted by Trifonov (2000), decreasing melting stability is accompanied by an increase in the complexity, for example the C-chain length, and a decrease in the chemical stability, of the encoded amino acids. Finally chain termination is directed by the least stable steps and does not require base pairing.
This analysis of the standard code neglects the phenomenon of coding variability where the same codon can encode different aminoacids in different organisms. This important aspect of the genetic code has been comprehensively reviewed by Osawa et al. (1992). In general the greatest differences in coding capacity occur between the standard code and non-plant mitochondria while significant, although less extensive differences are found between the standard code and that utilised in ciliates and certain Mycoplasma species. Strikingly, when the variability of coding capacity for particular codons is compared with the stability of the first base step the most extensive variation is associated entirely with the five least stable base steps (Table I). Notably some of the least stable triplets exhibit the greatest variability. For example in different contexts TGA can encode termination, cysteine, tryptophan or selenocysteine while TAA can encode termination, tyrosine or glutamine. In contrast the coding capacities of the five most stable base steps encoding alanine, glycine, valine, proline, glutamic acid, aspartic acid and arginine are essentially invariant.
A Model for the Evolution of the Genetic Code
These correlations described above are consistent with a simple model for the evolution of the genetic code (Figure 1). As suggested by both Jukes (1967) and Crick (1968), the initial code would have been a 2-letter triplet of the form XYNXYN... in which the codon–anticodon interactions would have been mediated by the XY base step. A coding box XYN would on this model specify a single amino acid. Subsequently a 3-letter triplet utilising the wobble rules would have developed generating a code of the form XYZXYZ... so that a coding box could specify two or more amino acids. Interestingly the CT/AG base step, which has an intermediate melting energy specifies one coding box with a single amino acid and a second with two (Table I). In this scenario the specification of an amino acid by a particular codon would depend largely on the availability of the cognate amino acid, accounting for the inverse correlation of base step stability with the complexity and chemical instability of the encoded amino acids. Further the conservation of coding capacity for the most stable base steps would be consistent with the view that codes which differ significantly from the standard code could have arisen by divergent evolution at a stage when only a few amino acids were specified. A similar explanation for the codes utilised by Mycoplasma, ciliates and some mitochondria arose from an early code prone to variability has been promulgated by Grivell (1986) and by Lewin (1990). The alternative view that non-standard codes arose entirely by evolution from a fixed standard code (Osawa et al. 1992) is not supported by the observed pattern of variability. Nevertheless the adoption of a novel coding capacity from the standard code would be consistent in some examples with the notion that the lower the energy of the codon–anticodon interaction the greater the possibility that this coding interaction could be usurped by an interloper. In the context of this discussion the mode of acquisition of the methionine codon(s) is uncertain. Certainly codon capture, either by isoleucine or methionine, is plausible.
This view of the evolution of the code raises the question as to why the code in its initial form should have been of the 2-letter triplet form rather than of a 2-letter doublet form. This issue was addressed by Crick (1968) who pointed out that if an initial RNA adaptor had a stem loop structure with the anti-codon on the loop, then the width of the stem would be ∼20 Å. In A-RNA the P-P distance ∼6 Å. The binding of two adaptors to successive codons requires contiguity of both codons and aminoacid linkage sites. Consider two types of doublet code: an uninterrupted form XYXYXY... and an interrupted form XYNXYNXYN. For a doublet interaction of XYXY type distance between successive codons is \( \sim 6 + 8 - 9 = 14 - 15 \) Å; for a doublet interaction of XYNXYN type distance between successive codons is \( \tilde\quad{}6 + 2{\left( {8 - 9} \right)} = 22 - 24 \) Å. Only in XYN case is there sufficient distance in mRNA to accommodate simultaneous binding of two adaptors – unless there is a sharp change of direction in polynucleotide chain which could abrogate contiguity of amino acid sites. This problem would be even more acute if there were originally three adaptors lined up, as in present day ribosomes. Even if the primordial mRNA were aligned on a positively charged surface a code of the form XYXYXY... would still be excluded.
One of the more striking features of the relationship between doublet base-pairing stability and coding capacity is the encoding of translation termination by the least stable base steps, TA and TG/CA. This correlation is fully consistent with an initial 2-letter triplet code. The complements TTA, CTA and TCA of the termination triplets TAA, TAG and TGA would in principle as a duplex have the same melting energies. Yet all encode an amino acid and in two of the three cases this amino acid is the sole representative in a coding box. This difference argues strongly that the first two bases in a codon, at least initially, determined the stability of a codon–anticodon interaction. If translation termination occurred by default where a stable codon–anticodon pairing could not be established then the weakest interactors would, again by default, remain as termination codons and would only be fixed in a stabilised genetic code when the specifying interaction was sufficiently energetically favourable. The concept that translation termination can be essentially a default mechanism in the absence of amino acid coding capacity is supported by the observations that the acquisition of coding capacity by mutation of a coding tRNA can suppress chain termination (Goodman et al. 1968) and that the termination codon TGA can encode the incorporation of selenocysteine provided that there is an additional signal to stabilise the codon–anticodon interaction (Zinoni et al. 1990; Berry et al. 1991).
The criterion of variability of coding capacity correlates extremely well with the stability of the interaction of the initial base step in a codon–anticodon pair. With one exception this correlation is paralleled by a unique amino acid assignment for each coding box. The exception is the GAN box which encodes both aspartic acid (GAY) and glutamic acid (GAR). Notably this pair of amino acids are also the most chemically similar in any of the coding boxes. Both are significant products of the early experiments to imitate a prebiotic environment (Miller and Urey 1959; Fox and Windsor 1970). Although a code of the form (XYN)n has, in principle, the capacity to specify only one entity, the entity specified does not have to be unique. Put another way, in terms of the initial specification of amino acids, the code could be sloppy with all the GAN codons directing the incorporation of either aspartic acid or glutamic acid. Only subsequently, possibly with the evolution of the ‘wobble’ rules, would discrimination between these two amino acids have evolved.
The ordering of amino acid coding capacity with base step stability differs from the consensus temporal order of codon acquisition deduced by Trifonov (2000, 2004) only in one, possibly minor, respect. This is relative position of arginine. The argument for a more ancient origin of the CGN set of arginine codons than is normally proposed is supported by three criteria. First the CG base step is predicted to be relatively stable, both by the Protozanova et al. (2004) and the Xia et al. (1998) data. Second, the CGN box codes only for a single amino acid, and finally the coding capacity is invariant. All these characteristics would be consistent with the CGN set of codons belonging originally to an (XYN)n code. However, arginine was not observed as a product of the electric discharge experiments of Miller (1953) and Miller and Urey (1959) although some was produced, but not quantitiated, when formaldehyde and ammonia were heated in the presence of silica (Fox and Windsor 1970). The failure to observe significant amounts of arginine in these experiments accounts in part for the difference in ordering between this discussion and that of Trifonov (2000). The question then becomes whether the early prebiotic synthesis of arginine is chemically plausible. This amino acid has an unbranched 5-carbon chain linked to a guanidinium group. Interestingly a major product of the Miller spark experiments, only exceeded in abundance by glycine and alanine, was the unbranched 5-carbon amino acid norleucine (Miller and Urey 1959). The generation of a guanidinium group could be facilitated by effective concentrations of ammonia and cyanide derivatives such as cyanamide and dicyanamide. Both hydrogen cyanide and dicyanamide have been invoked as significant primordial reactants for the production of biological precursor molecules (Basile et al. 1984; Steinman and Capolupo 1969). From this perspective two possible scenarios can be invoked: either the primordial concentration of arginine was sufficient for it initially to be encoded as such or, possibly less likely, CGN first coded for a different 5-carbon amino acid such as norleucine and subsequently this amino acid was replaced by arginine.
Conclusions
Despite a number of caveats the present day pattern of codon utilisation argues strongly that the energetics of codon–anticodon interactions were a determinative influence on the evolution of the genetic code and that any changes in the future are likely to be manifested as alterations in the specification by the least thermally stable codons.
References
Basile B, Lazcano A, Oro J (1984) Prebiotic syntheses of purines and pyrimidines. Adv Space Res 4:125–131
Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353:273–276
Crick FHC (1966) Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 19:548–555
Crick FHC (1968) The origin of the genetic code. J Mol Biol 38:367–379
Eigen M, Schuster P (1978) The hypercycle. A principle of natural self-organization. Part C. The realistic hypercycle. Naturwissenschafen 65:341–369
Fox SW, Windsor CR (1970) Synthesis of amino acids by heating formaldehyde and ammonia. Science 170:984–986
Goodman HM, Abelson J, Landy A, Brenner S, Smith JD (1968) Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature 217:1019–1024
Grivell LA (1986) Deciphering divergent codes. Nature 324:109–110
Hartman H (1975) Speculations on the evolution of the genetic code. Orig Life 6:423–427
Jukes TH (1967) Indications of an evolutionary pathway in the amino acid code. Biochem Biophys Res Commun 27:573–578
Lewin B (1990) Genes IV. Oxford University Press, Oxford (p 148)
Miller SL (1953) A production of amino acids under possible primitive earth conditions. Science 117:528–529
Miller SL, Urey HC (1959) Organic compound synthesis on the primitive earth. Science 130:245–251
Murphy FV 4th, Ramakrishnan V (2004) Structure of a purine–purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11:1251–1252
Osawa S, Jukes TH, Watanabe K, Muto A (1992) Recent evidence for evolution of the genetic code. Microbiol Rev 56:229–264
Protozanova E, Yakovchuk P, Frank-Kamenetskii MD (2004) Stacked-unstacked equilibrium at the nick site of DNA. J Mol Biol 342:775–785
SantaLucia J Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA 95:1460–1465
Steinman G, Capolupo AA (1969) The chemical plausibility of dicyanamide as a primordial reactant. Curr Mod Biol 2:295–297
Travers A (2007) DNA flexibility. Interface (in press)
Trifonov EN (2000) Consensus temporal order of amino acids and the evolution of the genetic code. Gene 261:139–151
Trifonov EN (2004) The triplet code from first principles. J Biomol Struct Dyn 22:1–11
Xia T, SantaLucia J Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37:14719–14735
Zinoni F, Heider J, Bock A (1990) Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci USA 87:4660–4664
Acknowledgements
I am most grateful to Dr. M. S. Bretscher for introducing me to this problem.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Travers, A. The Evolution of the Genetic Code Revisited. Orig Life Evol Biosph 36, 549–555 (2006). https://doi.org/10.1007/s11084-006-9041-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-006-9041-6