Abstract
Purpose
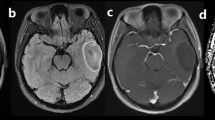
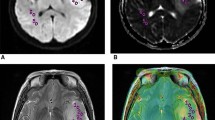
The prognosis of lower grade glioma (LGG) patients depends (in large part) on both isocitrate dehydrogenase (IDH) gene mutation and chromosome 1p/19q codeletion status. IDH-mutant LGG without 1p/19q codeletion (IDHmut-Noncodel) often exhibit a unique imaging appearance that includes high apparent diffusion coefficient (ADC) values not observed in other subtypes. The purpose of this study was to develop an ADC analysis-based approach that can automatically identify IDHmut-Noncodel LGG.
Methods
Whole-tumor ADC metrics, including fractional tumor volume with ADC > 1.5 × 10−3mm2/s (VADC>1.5), were used to identify IDHmut-Noncodel LGG in a cohort of N = 134 patients. Optimal threshold values determined in this dataset were then validated using an external dataset containing N = 93 cases collected from The Cancer Imaging Archive. Classifications were also compared with radiologist-identified T2-FLAIR mismatch sign and evaluated concurrently to identify added value from a combined approach.
Results
VADC>1.5 classified IDHmut-Noncodel LGG in the internal cohort with an area under the curve (AUC) of 0.80. An optimal threshold value of 0.35 led to sensitivity/specificity = 0.57/0.93. Classification performance was similar in the validation cohort, with VADC>1.5 ≥ 0.35 achieving sensitivity/specificity = 0.57/0.91 (AUC = 0.81). Across both groups, 37 cases exhibited positive T2-FLAIR mismatch sign—all of which were IDHmut-Noncodel. Of these, 32/37 (86%) also exhibited VADC>1.5 ≥ 0.35, as did 23 additional IDHmut-Noncodel cases which were negative for T2-FLAIR mismatch sign.
Conclusion
Tumor subregions with high ADC were a robust indicator of IDHmut-Noncodel LGG, with VADC>1.5 achieving > 90% classification specificity in both internal and validation cohorts. VADC>1.5 exhibited strong concordance with the T2-FLAIR mismatch sign and the combination of both parameters improved sensitivity in detecting IDHmut-Noncodel LGG.





Similar content being viewed by others
References
Network TCGAR (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498. https://doi.org/10.1056/NEJMoa1402121
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66(20):9852–9856
Smits M, van den Bent MJ (2017) Imaging correlates of adult glioma genotypes. Radiology 284(2):316–331. https://doi.org/10.1148/radiol.2017151930
Gutman DA, Cooper LAD, Hwang SN, Holder CA, Gao JJ, Aurora TD et al (2013) MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267(2):560–569
Patel SH, Poisson LM, Brat DJ, Zhou Y, Cooper L, Snuderl M et al (2017) T2–FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res 23(20):6078–6086
Wijnenga MMJ, French PJ, Dubbink HJ, Dinjens WNM, Atmodimedjo PN, Kros JM et al (2018) The impact of surgery in molecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro Oncol 20(1):103–112
Kawaguchi T, Sonoda Y, Shibahara I, Saito R, Kanamori M, Kumabe T et al (2016) Impact of gross total resection in patients with WHO grade III glioma harboring the IDH 1/2 mutation without the 1p/19q co-deletion. J Neurooncol 129(3):505–514
Patel SH, Bansal AG, Young EB, Batchala PP, Patrie JT, Lopes MB et al (2019) Extent of surgical resection in lower-grade gliomas: differential impact based on molecular subtype. Am J Neuroradiol 40(7):1149–1155
Foltyn M, Nieto Taborda KN, Neuberger U, Brugnara G, Reinhardt A, Stichel D et al (2020) T2/FLAIR-mismatch sign for noninvasive detection of IDH-mutant 1p/19q non-codeleted gliomas: validity and pathophysiology. Neuro-Oncol Adv 2(1):1–9
Broen MPG, Smits M, Wijnenga MMJ, Dubbink HJ, Anten MHME, Schijns OEMG et al (2018) The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncology 20(10):1393–1399
Batchala PP, Muttikkal TJE, Donahue JH, Patrie JT, Schiff D, Fadul CE et al (2019) Neuroimaging-based classification algorithm for predicting 1p/19q-codeletion status in IDH-mutant lower grade gliomas. Am J Neuroradiol 40(3):426–432
Jain R, Johnson DR, Patel SH, Castillo M, Smits M, van den Bent MJ et al (2020) “Real world” use of a highly reliable imaging sign: “T2-FLAIR mismatch” for identification of IDH mutant astrocytomas. Neuro-Oncology. https://doi.org/10.1093/neuonc/noaa041/5737809
Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ (2019) Imaging prediction of isocitrate dehydrogenase (IDH) mutation in patients with glioma: a systemic review and meta-analysis. Eur Radiol 29(2):745–758
Leu K, Ott GA, Lai A, Nghiemphu PL, Pope WB, Yong WH et al (2017) Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II–III diffuse gliomas. J Neurooncol 134(1):177–188
Xing Z, Yang X, She D, Lin Y, Zhang Y, Cao D (2017) Noninvasive assessment of IDH mutational status in World Health Organization Grade II and III astrocytomas using DWI and DSC-PWI combined with conventional MR imaging. Am J Neuroradiol 38(6):1138–1144
Lee S, Choi SH, Ryoo I, Yoon TJ, Kim TM, Lee S-H et al (2015) Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J Neurooncol 121(1):141–150
Wasserman JK, Nicholas G, Yaworski R, Wasserman A-M, Woulfe JM, Jansen GH et al (2015) Radiological and pathological features associated with IDH1-R132H mutation status and early mortality in newly diagnosed anaplastic astrocytic tumours. PLoS ONE 10(4):e0123890. https://doi.org/10.1371/journal.pone.0123890
Thust SC, Hassanein S, Bisdas S, Rees JH, Hyare H, Maynard JA et al (2018) Apparent diffusion coefficient for molecular subtyping of non-gadolinium-enhancing WHO grade II/III glioma: volumetric segmentation versus two-dimensional region of interest analysis. Eur Radiol 28(9):3779–3788
Aliotta E, Nourzadeh H, Batchala PP, Schiff D, Lopes MB, Druzgal JT et al (2019) Molecular subtype classification in lower-grade glioma with accelerated DTI. Am J Neuroradiol 40(9):1458–1463. https://doi.org/10.3174/ajnr.A6162
Wu C-C, Jain R, Radmanesh A, Poisson LM, Guo W-Y, Zagzag D et al (2018) Predicting genotype and survival in glioma using standard clinical MR imaging apparent diffusion coefficient images: a pilot study from the cancer genome atlas. AJNR Am J Neuroradiol 39(10):1814–1820
Xiong J, Tan W, Wen J, Pan J, Wang Y, Zhang J et al (2016) Combination of diffusion tensor imaging and conventional MRI correlates with isocitrate dehydrogenase 1/2 mutations but not 1p/19q genotyping in oligodendroglial tumours. Eur Radiol 26(6):1705–1715. https://doi.org/10.1007/s00330-015-4025-4
Fellah S, Caudal D, De Paula AM, Dory-Lautrec P, Figarella-Branger D, Chinot O et al (2013) Multimodal MR imaging (diffusion, perfusion, and spectroscopy): is it possible to distinguish oligodendroglial tumor grade and 1p/19q codeletion in the pretherapeutic diagnosis? Am J Neuroradiol 34(7):1326–1333
Park YW, Han K, Ahn SS, Choi YS, Chang JH, Kim SH et al (2018) Whole-tumor histogram and texture analyses of DTI for evaluation of IDH1-mutation and 1p/19q-codeletion status in world health organization grade II gliomas. Am J Neuroradiol 39(4):693–698
Feng X, Tustison N, Meyer C (2019) Brain tumor segmentation using an ensemble of 3D U-nets and overall survival prediction using radiomic features. Lect Notes Comput Sci 11384 LNCS(April):279–288
Capper D, Weißert S, Balss J, Habel A, Meyer J, Jãger D et al (2010) Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol 20(1):245–254
Felsberg J, Wolter M, Seul H, Friedensdorf B, Göppert M, Sabel MC et al (2010) Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol 119(4):501–507
Riemenschneider MJ, Jeuken JWM, Wesseling P, Reifenberger G (2010) Molecular diagnostics of gliomas: state of the art. Acta Neuropathol 120(5):567–584
Foster JM, Oumie A, Togneri FS, Vasques FR, Hau D, Taylor M et al (2015) Cross-laboratory validation of the OncoScan® FFPE Assay, a multiplex tool for whole genome tumour profiling. BMC Med Genom 8(1):5
The Cancer Genome Atlas - LGG Collection. https://wiki.cancerimagingarchive.net/display/Public/TCGA-LGG#6abaca285cee4c9cac59b0bcff944658. Accessed 1 Mar 2020
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P et al (2013) The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J et al (2017) Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection [Data Set]. The Cancer Imaging Archive. https://wiki.cancerimagingarchive.net/display/DOI/Segmentation+Labels+and+Radiomic+Features+for+the+Pre-operative+Scans+of+the+TCGA-LGG+collection
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS et al (2017) Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 4(March):1–13
The Cancer Genome Atlas Data Matrix. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Accessed 4 May 2020
Bakas S, Zeng K, Sotiras A, Rathore S, Akbari H, Gaonkar B, et al (2016) GLISTRboost: combining multimodal MRI segmentation, registration, and biophysical tumor growth modeling with gradient boosting machines for glioma segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer, Berlin, pp 144–155
Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA et al (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164(3):550–563
Li DL, Shen F, Yin Y, Peng JX, Chen PY (2013) Weighted youden index and its two-independent-sample comparison based on weighted sensitivity and specificity. Chin Med J 126(6):1150–1154
Corell A, Ferreyra Vega S, Hoefling N, Carstam L, Smits A, Olsson Bontell T et al (2020) The clinical significance of the T2-FLAIR mismatch sign in grade II and III gliomas: a population-based study. BMC Cancer. https://doi.org/10.1186/s12885-020-06951-w
Lasocki A, Tsui A, Gaillard F, Tacey M, Drummond K, Stuckey S (2019) Reliability of noncontrast-enhancing tumor as a biomarker of IDH1 mutation status in glioblastoma. J Clin Neurosci 39:170–175
Nakae S, Murayama K, Sasaki H, Kumon M, Nishiyama Y, Ohba S et al (2017) Prediction of genetic subgroups in adult supra tentorial gliomas by pre- and intraoperative parameters. J Neurooncol 131(2):403–412
Qi S, Yu L, Li H, Ou Y, Qiu X, Ding Y et al (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 7(6):1895–1902
Delfanti RL, Piccioni DE, Handwerker J, Bahrami N, Krishnan A, Karunamuni R et al (2017) Imaging correlates for the 2016 update on WHO classification of grade II/III gliomas: implications for IDH, 1p/19q and ATRX status. J Neurooncol 135(3):601–609
Saito T, Muragaki Y, Maruyama T, Komori T, Tamura M, Nitta M et al (2016) Calcification on CT is a simple and valuable preoperative indicator of 1p/19q loss of heterozygosity in supratentorial brain tumors that are suspected grade II and III gliomas. Brain Tumor Pathol 33(3):175–182. https://doi.org/10.1007/s10014-016-0249-5
Patel SH, Batchala PP, Mrachek EKS, Lopes M-BS, Schiff D, Fadul CE et al (2020) MRI and CT identify isocitrate dehydrogenase (IDH)-mutant lower-grade gliomas misclassified to 1p/19q codeletion status with fluorescence in situ hybridization. Radiology 294(1):160–167
van der Voort SR, Incekara F, Wijnenga MMJ, Kapas G, Gardeniers M, Schouten JW et al (2019) Predicting the 1p/19q codeletion status of presumed low-grade glioma with an externally validated machine learning algorithm. Clin Cancer Res 25(24):7455–7462
Akkus Z, Ali I, Sedlář J, Agrawal JP, Parney IF, Giannini C et al (2017) Predicting deletion of chromosomal arms 1p/19q in low-grade gliomas from MR images using machine intelligence. J Digit Imaging 30(4):469–476
Li Z, Wang Y, Yu J, Guo Y, Cao W (2017) Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep 7(1):1–11
Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G et al (2018) Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol 39(7):1201–1207
Han Y, Xie Z, Zang Y, Zhang S, Gu D, Zhou M et al (2018) Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J Neurooncol 140(2):297–306
Shboul ZA, Chen JM, Iftekharuddin K (2020) Prediction of molecular mutations in diffuse low-grade gliomas using MR imaging features. Sci Rep 10(1):1–13
Kim D, Wang N, Ravikumar V, Raghuram DR, Li J, Patel A et al (2019) Prediction of 1p/19q codeletion in diffuse glioma patients using pre-operative multiparametric magnetic resonance imaging. Front Comput Neurosci 13:52. https://doi.org/10.3389/fncom.2019.00052/full
Lu CF, Hsu FT, Hsieh KLC, Kao YCJ, Cheng SJ, Hsu JBK et al (2018) Machine learning–based radiomics for molecular subtyping of gliomas. Clin Cancer Res 24(18):4429–4436
Zhou H, Vallières M, Bai HX, Su C, Tang H, Oldridge D et al (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncology 19(6):862–870
Tan W, Xiong J, Huang W, Wu J, Zhan S, Geng D (2017) Noninvasively detecting Isocitrate dehydrogenase 1 gene status in astrocytoma by dynamic susceptibility contrast MRI. J Magn Reson Imaging 45(2):492–499
Yamashita K, Hiwatashi A, Togao O, Kikuchi K, Hatae R, Yoshimoto K et al (2016) MR imaging–based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. Am J Neuroradiol 37(1):58–65
Choi C, Raisanen JM, Ganji SK, Zhang S, McNeil SS, An Z et al (2016) Prospective longitudinal analysis of 2-hydroxyglutarate magnetic resonance spectroscopy identifies broad clinical utility for the management of patients with IDH-mutant glioma. J Clin Oncol 34(33):4030–4039
Tietze A, Choi C, Mickey B, Maher EA, Parm Ulhøi B, Sangill R et al (2018) Noninvasive assessment of isocitrate dehydrogenase mutation status in cerebral gliomas by magnetic resonance spectroscopy in a clinical setting. J Neurosurg 128(2):391–398
Lee MK, Park JE, Jo Y, Park SY, Kim SJ, Kim HS (2020) Advanced imaging parameters improve the prediction of diffuse lower-grade gliomas subtype, IDH mutant with no 1p19q codeletion: added value to the T2/FLAIR mismatch sign. Eur Radiol 30(2):844–854
Funding
Grant Support: S.H.P.: RSNA Research Scholar Grant (RSCH1819).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aliotta, E., Dutta, S.W., Feng, X. et al. Automated apparent diffusion coefficient analysis for genotype prediction in lower grade glioma: association with the T2-FLAIR mismatch sign. J Neurooncol 149, 325–335 (2020). https://doi.org/10.1007/s11060-020-03611-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-020-03611-8