Abstract
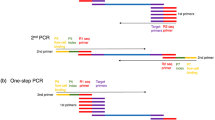
As more and more re-sequencing genome data in crops were released, SNPs (single nucleotide polymorphisms) are easily achieved for genotyping and show the highest abundance among all kinds of molecular markers. However, high-throughput SNP genotyping methods, such as KASP (Kompetitive Allele Specific PCR), TaqMan, or ARMS (amplification refractory mutation system), are always labor on DNA extraction and depending on expensive equipment. Therefore, most breeders are undergoing the bottleneck of lacking an easy, cost-saving, and stable genotyping system. Here, we report a direct PCR–based medium-throughput SNP marker–assisted selection (D-MAS) system suits available major crops including rice, wheat, maize, and rapeseed. The D-MAS system, which reduces the time of manual operation and result analysis dramatically, contains (1) seedling breeding in greenhouse; (2) high-throughput DNA extraction by alkaline lysis; and (3) gel-free SNP marker detection with 384-well by PARMS (penta-primer amplification refractory mutation) or KASP genotyping system. The stability of alkaline lysis DNA was validated by flexible dilution fold and long storing time under low temperatures. The alkaline lysis DNA from four F2 populations of rice, wheat, maize, and rapeseed showed equal efficiency in SNP calling relative to the ones from cetyl trimethylammonium bromide (CTAB) method. With the alkaline lysis DNA, PAMRS showed denser genotype cluster than KASP. Furthermore, the D-MAS system was adaptive with rice old leaves. The throughput and efficiency of the D-MAS system were validated in the hybrid rice seed purity test with a Xian-Geng-specific SNP marker which is also recommended to select ipa1-2d gene in the rice molecular breeding. In conclusion, we proposed a direct PCR–based SNP calling pipeline, which could be a simple, cheap, and robust standard operation procedure (SOP) of molecular breeding for different crops and get extensive use in most laboratory.




Similar content being viewed by others
References
Batley J (2015) Plant genotyping: methods and protocols. In: Methods in Molecular Biology, vol 1245. Springer, New York
Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, Jenkins J, Barry K, Lindquist E, Hellsten U, Deshpande S, Wang X, Wu X, Mitros T, Triplett J, Yang X, Ye CY, Mauro-Herrera M, Wang L, Li P, Sharma M, Sharma R, Ronald PC, Panaud O, Kellogg EA, Brutnell TP, Doust AN, Tuskan GA, Rokhsar D, Devos KM (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30(6):555–561. https://doi.org/10.1038/nbt.2196
Bukowski R, Guo X, Lu Y, Zou C, He B, Rong Z, Wang B, Xu D, Yang B, Xie C, Fan L, Gao S, Xu X, Zhang G, Li Y, Jiao Y, Doebley JF, Ross-Ibarra J, Lorant A, Buffalo V, Romay MC, Buckler ES, Ware D, Lai J, Sun Q, Xu Y (2018) Construction of the third-generation Zea mays haplotype map. GigaScience 7(4):1–12. https://doi.org/10.1093/gigascience/gix134
Dayteg C, Tuvesson S, Merker A, Jahoor A, Brantestam KA (2007) Automation of DNA marker analysis for molecular breeding in crops: practical experience of a plant breeding company. Plant Breed 126(4):410–415. https://doi.org/10.1111/j.1439-0523.2007.01306.x
Eathington SR, Crosbie TM, Edwards MD, Reiter RS, Bull JK (2007) Molecular markers in a commercial breeding program. Crop Sci 47(Supplement_3):S-154
Heim M, Meyer UA (1990) Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet 336(8714):529–532. https://doi.org/10.1016/0140-6736(90)92086-W
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42(11):961–967. https://doi.org/10.1038/ng.695
Huang X, Yang S, Gong J, Zhao Y, Feng Q, Gong H, Li W, Zhan Q, Cheng B, Xia J, Chen N, Hao Z, Liu K, Zhu C, Huang T, Zhao Q, Zhang L, Fan D, Zhou C, Lu Y, Weng Q, Wang ZX, Li J, Han B (2015) Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun 6:6258. https://doi.org/10.1038/ncomms7258
Huang X, Yang S, J-Y G, Zhao Q, Feng Q, Zhan Q, Zhao Y, Li W, Cheng B, Xia J, Chen N, Huang T, Zhang L, Fan D, Chen J, Zhou C, Lu Y, Weng Q, Han B (2016) Genomic architecture of heterosis for yield traits in rice. Nature 537. https://doi.org/10.1038/nature19760
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436(7052):793–800. https://doi.org/10.1038/nature03895
International Wheat Genome Seqencing Consortium(IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345(6194):1251788. https://doi.org/10.1126/science.1251788
Ma C, Ma X, Yao L, Liu Y, Du F, Yang X, Xu M (2017) qRfg3, a novel quantitative resistance locus against Gibberella stalk rot in maize. Theor Appl Genet 130(8):1723–1734. https://doi.org/10.1007/s00122-017-2921-5
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4325
Myakishev MV, Khripin Y, Hu S, Hamer DH (2001) High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res 11(1):163–169
Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17(7):2503–2516. https://doi.org/10.1093/nar/17.7.2503
Nuovo GJ, Hohman RJ, Nardone GA, Nazarenko IA (1999) In situ amplification using universal energy transfer-labeled primers. J Histochem Cytochem 47(3):273–279. https://doi.org/10.1177/002215549904700301
Pan Q, Xu Y, Li K, Peng Y, Zhan W, Li W, Li LAND, Yan J (2017) The genetic basis of plant architecture in 10 maize recombinant inbred line populations. Plant Physiol 175:858–873
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann M, Ming R, Peterson DG, Mehboob-ur-Rahman, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457(7229):551–556. https://doi.org/10.1038/nature07723
Rasheed A, Wen W, Gao F, Zhai S, Jin H, Liu J, Guo Q, Zhang Y, Dreisigacker S, Xia X, He Z (2016) Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor Appl Genet 129(10):1843–1860. https://doi.org/10.1007/s00122-016-2743-x
Rosas JE, Bonnecarrère V, Pérez de Vida F (2014) One-step, codominant detection of imidazolinone resistance mutations in weedy rice (Oryza sativa L.). Electron J Biotechnol 17:95–101
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463(7278):178–183. https://doi.org/10.1038/nature08670
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann M, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang L, Yu Y, Zhang L, Zhou S, Zhu Q, Bennetzen JL, Dawe RK, Jiang J, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie W, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. https://doi.org/10.1126/science.1178534
Thomson JM (2014) High-throughput SNP genotyping to accelerate crop improvement. Plant Breed Biotechnol 2(3):195–212
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Yu S (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44(10):1098–1103. https://doi.org/10.1038/ng.2371
Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu J, Sun C, Fu B, Zhang H, Gao Y, Zhao X, Shen F, Cui X, Yu H, Li Z, Chen M, Detras J, Zhou Y, Zhang X, Zhao Y, Kudrna D, Wang C, Li R, Jia B, Lu J, He X, Dong Z, Xu J, Li Y, Wang M, Shi J, Li J, Zhang D, Lee S, Hu W, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu Y, Yue Z, Naredo MEB, Talag J, Wang X, Li J, Fang X, Yin Y, Glaszmann JC, Zhang J, Li J, Hamilton RS, Wing RA, Ruan J, Zhang G, Wei C, Alexandrov N, McNally K, Li Z, Leung H (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557(7703):43–49. https://doi.org/10.1038/s41586-018-0063-9
Wing RA, Purugganan MD, Zhang Q (2018) The rice genome revolution: from an ancient grain to Green Super Rice. Nat Rev Genet 19(8):505–517. https://doi.org/10.1038/s41576-018-0024-z
Yang G, Chen S, Chen L, Sun K, Huang C, Zhou D, Huang Y, Wang J, Liu Y, Wang H (2019) Development of a core SNP arrays based on the KASP method for molecular breeding of rice. Rice 12:21. https://doi.org/10.1186/s12284-019-0272-3
Zeng D, Tian Z, Rao Y, Dong G, Yang Y, Huang L, Leng Y, Xu J, Sun C, Zhang G, Hu J, Zhu L, Gao Z, Hu X, Guo L, Xiong G, Wang Y, Li J, Qian Q (2017) Rational design of high-yield and superior-quality rice. Nat Plants 3:17031. https://doi.org/10.1038/nplants.2017.31
Zhang J, Chen L-L, Xing F, Kudrna DA, Yao W, Copetti D, Mu T, Li W, Song J-M, Xie W, Lee S, Talag J, Shao L, An Y, Zhang C-L, Ouyang Y, Sun S, Jiao W-B, Lv F, Du B, Luo M, Maldonado CE, Goicoechea JL, Xiong L, Wu C, Xing Y, Zhou D-X, Yu S, Zhao Y, Wang G, Yu Y, Luo Y, Zhou Z-W, Hurtado BEP, Danowitz A, Wing RA, Zhang Q (2016) Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc Natl Acad Sci U S A 113(35):E5163–E5171. https://doi.org/10.1073/pnas.1611012113
Zhang L, Yu H, Ma B, Liu G, Wang J, Gao R, Li J, Liu J, Xu J, Zhang Y, Li Q, Huang X, Qian Q, Han B, He Z (2017) A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat Commun 8:14789. https://doi.org/10.1038/ncomms14789
Zhang Q, Li J, Xue Y, Han B, Deng XW (2008) Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol Plant 1(5):715–719. https://doi.org/10.1093/mp/ssn043
Zhao J, Wang Z, Liu H, Li T, Hou J, Zhang X, Hao C (2019) Global status of 47 major wheat loci controlling yield, quality, adaptation and stress resistance selected over the last century. BMC Plant Biol 19(1):5. https://doi.org/10.1186/s12870-018-1612-y
Acknowledgments
We thank Dr. Haitao Li, Dr. YiKe Liu, and Mr. Duo Wang for their kindly providing rapeseed, wheat, and maize F2 population. We gratefully acknowledge Dr. Huihui Yu for the advice and revision of this manuscript.
Funding information
This research was supported by the Fundamental Research Funds for the Central Universities of China (Grant No. 2662017QD033), Hubei Provincial Natural Science Foundation of China (Grant No. 2019CFA061), Special Major Projects for Technological Innovation in Hubei Province (2019ABA104), National Natural Science Foundation of China (Grant No.31600983), and the Earmarked Fund for the China Agriculture Research System of China (Grant No. CARS-01-05).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Lu, J., Hou, J., Ouyang, Y. et al. A direct PCR–based SNP marker–assisted selection system (D-MAS) for different crops. Mol Breeding 40, 9 (2020). https://doi.org/10.1007/s11032-019-1091-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1091-3