Abstract
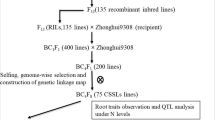
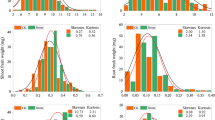
Aluminum (Al) toxicity is a major constraint on crop production in acid soils around the world. Hexaploid oat (Avena sativa L.) possesses significant Al tolerance making it a good candidate for production in these environments. Genetic improvement for Al tolerance in oat has traditionally been achieved through conventional plant breeding and could be enhanced by marker-assisted selection. The objectives of this study were to develop a chromosome-anchored genetic map for an oat recombinant inbred population and to identify SNP markers linked to quantitative trait loci (QTL) affecting root growth response to Al. Three QTL on chromosomes 7C-17A, 13A, and 19A conferring Al tolerance were identified using primary root regrowth of recombinant inbred lines derived from the cross between UFRGS 17 (Al tolerant) and UFRGS 930598-6 (Al sensitive). Localization of each QTL onto the sequenced rice genome revealed the genetic region on chromosome 13A might be associated with a putative malate transporter locus (LOC_Os06g15779). Studies of root apex tissue indicated that exudation of malate was increased in the Al-tolerant parent UFRGS17 and not in the sensitive parent. Based on these data, the malate transporter might be a candidate gene responsible for one of the Al tolerance QTL identified in this study.


Similar content being viewed by others
References
Archambault DJ, Zhang GC, Taylor GJ (1997) Spatial variation in the kinetics of aluminum (Al) uptake in roots of wheat (Triticum aestivum L.) exhibiting differential resistance to Al—evidence for metabolism-dependent exclusion of Al. J Plant Physiol 151:668–674
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–219
Basu U, Good AG, Taing-Aung T, Slaski J, Basu A, Briggs KG, Taylor GJ (1999) A 23 kDa protein root exudates polypeptide co-segregates with aluminium resistance in Triticum aestivum. Physiol Plant 106:53–61
Bian M et al (2013) Development of gene-specific markers for acid soil/aluminium tolerance in barley (Hordeum vulgare L.). Mol Breed 32:155–164. doi:10.1007/s11032-013-9859-3
Bianchi-Hall CM et al (2000) Aluminum tolerance associated with quantitative trait loci derived from soybean PI 416937 in hydroponics. Crop Sci 40:538–545. doi:10.2135/cropsci2000.402538x
Bryan GJ, Stephenson P, Collins A, Kirby J, Smith JB, Gale MD (1999) Low levels of DNA sequence variation among adapted genotypes of hexaploid wheat. Theor Appl Genet 99:192–198. doi:10.1007/s001220051224
Castilhos G, Farias JG, de Bernardi Schneider A, de Oliveira PH, Nicoloso FT, Chitolina Schetinger MR, Delatorre CA (2011) Aluminum-stress response in oat genotypes with monogenic tolerance. Environ Exp Bot 74:114–121. doi:10.1016/j.envexpbot.2011.05.007
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Collins NC, Shirley NJ, Saeed M, Pallotta M, Gustafson JP (2008) An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 179:669–682. doi:10.1534/genetics.107.083451
Consortium TRCS (2005) Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res 15:1284–1291. doi:10.1101/gr.3869505
Delhaize E, Gruber BD, Ryan PR (2007) The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett 581:2255–2262. doi:10.1016/j.febslet.2007.03.057
Foy CD, Chaney RL, White MC (1978) The physiology of metal toxicity in plants. Annu Rev Plant Physiol 29:511–566. doi:10.1146/annurev.pp.29.060178.002455
Fujii M et al (2012) Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun 3:713. http://www.nature.com/ncomms/journal/v3/n3/suppinfo/ncomms1726_S1.html
Genome sequencing and analysis of the model grass Brachypodium distachyon (2010). Nature 463:763–768. http://www.nature.com/nature/journal/v463/n7282/suppinfo/nature08747_S1.html
Gustafsson JP (2010) Visual MINTEQ ver. 3.0
Hervé CB, Calai FA, Nava IC, Delatorre CA (2013) Tolerância ao alumínio tóxico em germoplasma brasileiro elite de aveia. Ciência Rural 43:1364–1370
Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21:655–667. doi:10.1105/tpc.108.064543
Huan-Xin J, Ning T, Jin-Gui Z, Yan L, Li-Song C (2009) Phosphorus alleviates aluminum-induced inhibition of growth and photosynthesis in Citrus grandis seedlings. Physiol Plant 137:298–311
Jellen E, Beard J (2000) Geographical distribution of a chromosome 7C and 17 intergenomic translocation in cultivated oat. Crop Sci 40:256–263
Jellen EN, Gill BS, Cox TS (1994) Genomic in situ hybridization differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena). Genome 37:613–618. doi:10.1139/g94-087
Kobayashi Y et al (2013) Molecular and physiological analysis of Al3+ and H+ rhizotoxicities at moderately acidic conditions. Plant Physiol 163:180–192. doi:10.1104/pp.113.222893
Kochian LV, Piñeros MA, Hoekenga OA (2005) Physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274:175–195
Kulcheski FR, Graichen FAS, Martinelli JA, Locatelli AB, Federizzi LC, Delatorre CA (2010) Molecular mapping of Pc68, a crown rust resistance gene in Avena sativa. Euphytica 175:423–432
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li CD, Rossnagel BG, Scoles GJ (2000) The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor Appl Genet 101:1259–1268. doi:10.1007/s001220051605
Liu J, Magalhães JV, Shaff J, Kochian LV (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. doi:10.1111/j.1365-313X.2008.03696.x
Ma H-X, Bai G-H, Lu W-Z (2006) Quantitative trait loci for aluminum resistance in wheat cultivar Chinese Spring. Plant Soil 283:239–249. doi:10.1007/s11104-006-0008-1
Magalhães JV et al (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminium tolerance in sorghum. Nat Genet 39:1156–1161
Maron LG et al (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61:728–740. doi:10.1111/j.1365-313X.2009.04103.x
Merino-Gergichevic C, Alberdi M, Ivanov AG, Reyes-Diaz M (2010) Al+3–Ca+2 interaction in plants growing in acid soils: Al-phytotoxicity response to calcareous amendments. J Soil Sci Plant Nutr 10:217–243
Minella E, Sorrells ME (1997) Inheritance and chromosome location of Alp, a gene controlling aluminum tolerance in ‘Dayton’ barley. Plant Breed 116:465–469. doi:10.1111/j.1439-0523.1997.tb01032.x
Murphy JP, Hoffman LA (1992) The origin, history, and production of oat. In: Marshall HG, Sorrells ME (eds) Oat science and technology. ASA, CSSA Publishers, Madison, pp 1–28
Nava IC, Delatorre CA, Duarte I, Pacheco MT, Federizzi LC (2006) Inheritance of aluminum tolerance and its effects on grain yield and grain quality in oats (Avena sativa L.). Euphytica 148:353–358
Nava IC, Delatorre CA, Pacheco MT, Scheeren PL, Federizzi LC (2015) Aluminum tolerance of oat cultivars under hydroponic and acid soil conditions. Exp Agric. doi:10.1017/S0014479715000046
Navakode S, Weidner A, Lohwasser U, Röder MS, Börner A (2009) Molecular mapping of quantitative trait loci (QTLs) controlling aluminium tolerance in bread wheat. Euphytica 166:283–290. doi:10.1007/s10681-008-9845-8
Nguyen VT, Burow MD, Nguyen HT, Le BT, Le TD, Paterson AH (2001) Molecular mapping of genes conferring aluminum tolerance in rice (Oryza sativa L.). Theor Appl Genet 102:1002–1010. doi:10.1007/s001220000472
O’Donoughue LS et al (1995) A molecular linkage map of cultivated oat. Genome 38:368–380. doi:10.1139/g95-048
Ofei-manu P, Wagatsuma T, Ishikawa S, Tawaraya K (2001) The plasma membrane strength of the root-tip cells ans root phenolic compounds are correlated with Al tolerance in several common woody plants. Soil Sci Plant Nutr 47:359–375
Oliveira PH, Federizzi LC, Milach SCK, Gotuzzo C, Sawasato JT (2005) Inheritance in oat (Avena sativa L.) of tolerance to soil aluminum toxicity. Crop Breed Appl Biotechnol 5:125–133
Oliver RE et al (2013) SNP discovery and chromosome anchoring provide the first physically-anchored hexaploid oat map and reveal synteny with model species. PLoS ONE 8:e58068. doi:10.1371/journal.pone.0058068
Pellet DM, Papernik LA, Kochian LV (1996) Multiple aluminum-resistance mechanisms in wheat: roles of root apical phosphate and malate exudation. Plant Physiol 112:591–597
Peng J, Ronin Y, Fahima T, Röder MS, Li Y, Nevo E, Korol A (2003) Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci 100:2489–2494. doi:10.1073/pnas.252763199
Pereira LB et al (2013) Differential speed of activation in antioxidant system in three oat genotypes. J Inorg Biochem 128:202–207. doi:10.1016/j.jinorgbio.2013.07.025
Portyanko VA, Hoffman DL, Lee M, Holland JB (2001) A linkage map of hexaploid oat based on grass anchor DNA clones and its relationship to other oat maps. Genome 44:249–265
Project RA (2008) The rice annotation project database (RAP-DB): 2008 update. Nucleic Acids Res 36:D1028–D1033. doi:10.1093/nar/gkm978
Radmer L, Tesfaye M, Somers D, Temple S, Vance C, Samac D (2012) Aluminum resistance mechanisms in oat (Avena sativa L.). Plant Soil 351:121–134. doi:10.1007/s11104-011-0937-1
Raman H et al (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791. doi:10.1139/g05-054
Rines H, Molnar S, Tinker N, Phillips R (2006) Oat. Genome mapping and molecular breeding in plants. Cereals Millets 1:211–242
Rossiello RO, Netto JJ (2006) Toxidez de alumínio em plantas: novos enfoques para um velho problema. In: Fernandes MS (ed) Nutrição mineral de plantas. Sociedade Brasileira de Ciência do Solo, Viçosa, pp 375–418
Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 62:9–20. doi:10.1093/jxb/erq272
Sanchez-Chacon CD, Federizzi LC, Milach SCK, Pacheco MT (2000) Viabilidade genética e herança da tolerância à toxicidade do aluminio em aveia. Pesqui Agropecu Bras 35:1797–1808
Sasaki T et al (2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol 47:1343–1354. doi:10.1093/pcp/pcl002
Silva-Navas J, Benito C, Téllez-Robledo B, Abd El-Moneim D, Gallego FJ (2012) The ScAACT1 gene at the Q alt5 locus as a candidate for increased aluminum tolerance in rye (Secale cereale L.). Mol Breed 30:845–856. doi:10.1007/s11032-011-9668-5
Sivaguru M et al (2000) Aluminum-induced 1-3-B-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol 2:991–1006
Sorrells ME et al (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827. doi:10.1101/gr.1113003
Soto-Cerda B, Peñaloza E, Montenegro A, Rupayan A, Gallardo M, Salvo-Garrido H (2013) An efficient marker-assisted backcrossing strategy for enhancing barley (Hordeum vulgare L.) production under acidity and aluminium toxicity. Mol Breed 31:855–866. doi:10.1007/s11032-013-9839-7
Tan YD, Wan C, Zhu Y, Lu C, Xiang Z, Deng HW (2001) An amplified fragment length polymorphism map of the silkworm. Genetics 157:1277–1284
Tinker NA, Mather DE (1995) MQTL: software for simplified composite interval mapping of QTL in multiple environments. J Agric Genomics 1:1–4
Tinker NA et al (2014) A SNP genotyping array for hexaploid oat. Plant Genome. doi:10.3835/plantgenome2014.03.0010
Vogl C, Xu S (2000) Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155:1439–1447
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wagner CW, Milach SCK, Federizzi LC (2001) Genetic inheritance of aluminum tolerance in oat. Crop Breed Appl Biotechnol 1:22–26
Wang J et al (2007) High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 115:265–276. doi:10.1007/s00122-007-0562-9
Wang J et al (2009) High-throughput single nucleotide polymorphism genotyping using nanofluidic Dynamic Arrays. BMC Genomics 10:561
Wight CP et al (2003) A molecular marker map in ‘Kanota’ x ‘Ogle’ hexaploid oat (Avena spp.) enhanced by additional markers and a robust framework. Genome 46:28–47
Yokosho K, Yamaji N, Ma JF (2011) An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J 68:1061–1069. doi:10.1111/j.1365-313X.2011.04757.x
Yu J, Herrmann M (2006) Inheritance and mapping of a powdery mildew resistance gene introgressed from Avena macrostachya in cultivated oat. Theor Appl Genet 25:329–335
Zheng SJ, Ma JF, Matsumoto H (1998) Continuous secretion of organic acids is related to aluminium resistance during relatively long-term exposure to aluminium stress. Physiol Plant 103:209–214
Zheng SJ, Yang JL, He YF, Yu XH et al (2005) Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol 138:297–303
Zhou G, Delhaize E, Zhou M, Ryan PR (2013) The barley MATE gene, HvAACT1, increases citrate efflux and Al3+ tolerance when expressed in wheat and barley. Ann Bot 112:603–612. doi:10.1093/aob/mct135
Acknowledgments
The authors thank Dr. Eric Jellen and Dr. Jeff Maughan from Brigham Young University for the support during the SNP analysis; Dr. Marcelo T. Pacheco for the heritability analysis, and Mauricio D. Salomon for figure preparation. This study was supported by grants, scholarships, and fellowships from the Brazilian National Council for Scientific and Technological Development (CNPq), the Coordination for the Improvement of Higher Education Personnel (CAPES) from Brazil, The Prairie Oat Growers Association, and the US Department of Agriculture.
Conflict of interest
The authors declare that they have no conflict of interest and the experiments complied with the current laws of Brazil and USA, where they were performed.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schneider, A.B., Nava, I.C., Hervé, C.B. et al. Chromosome-anchored QTL conferring aluminum tolerance in hexaploid oat. Mol Breeding 35, 121 (2015). https://doi.org/10.1007/s11032-015-0315-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0315-4