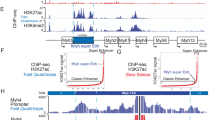
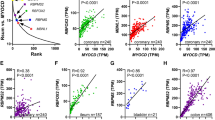
Summary
We have analyzed nearly 2,000 myosin heavy chain gene (Myh) clones representing over 30 different transcripts from seven of eight striated muscle Myh genes expressed in mouse. We also report the transcriptional start sites (TSS) for the mouse developmental Myh genes. The data reveal a previously unknown diversity of TSSs and 5′-end alternative splicing in these transcripts. The cardiac Myh6 gene had two major TSSs. Use of the major downstream site led to an alternatively spliced second exon. Each of the other Myh genes had one major TATA-directed TSS and one or more minor alternative TSSs, some associated with alternative splicing. The minor transcripts were associated with polysomes and their spatial-temporal expression largely mirrored that of the major transcripts in wild-type, Myh1 null, Myh4 null, injured, and uninjured muscle, except that one form of Myh7, detected in heart, was not detected in diaphragm, and the ratio of the two major Myh6 transcripts varied in some circumstances. These findings indicate that alternative TSS usage and alternative splicing in the 5′-UTR are a general feature of murine Myh gene expression and that Myh gene regulation is more complex than previously appreciated.
Similar content being viewed by others
References
Acakpo-Satchivi LJ, Edelmann W, Sartorius C, Lu BD, Wahr PA, Watkins SC, Metzger JM, Leinwand L, Kucherlapati R (1997) Growth and muscle defects in mice lacking adult myosin heavy chain genes. J Cell Biol 139:1219–1229
Allen DL, Sartorius CA, Sycuro LK, Leinwand LA (2001a) Different pathways regulate expression of the skeletal myosin heavy chain genes. J Biol Chem 276:43524–43533
Allen DL, Harrison BC, Sartorius C, Byrnes WC, Leinwand LA (2001b) Mutation of the IIB myosin heavy chain gene results in muscle fiber loss and compensatory hypertrophy. Am J␣Physiol Cell Physiol 280:C637–C645
Allen DL, Leinwand LA (2002) Intracellular calcium and myosin isoform transitions. Calcineurin and calcium-calmodulin kinase pathways regulate preferential activation of the IIa myosin heavy chain promoter. J Biol Chem 277:45323–45330
Allen DL, Weber JN, Sycuro LK, Leinwand LA (2005) Myocyte enhancer factor-2 and serum response factor binding elements regulate fast myosin heavy chain transcription in vivo. J Biol Chem 280:17126–17134
Babij P, Periasamy M (1989) Myosin heavy chain isoform diversity in smooth muscle is produced by differential RNA processing. J Mol Biol 210:673–679
Baldwin KM, Haddad F (2001) Plasticity in skeletal, cardiac and smooth muscle: effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J Appl Physiol 90:345–357
Beylkin DH, Allen DL, Leinwand LA (2006) MyoD, Myf5, and the calcineurin pathway activate the developmental myosin heavy chain genes. Dev Biol (in press). DOI 10.1016/j. ydbio.2006.02.04
Berne RM, Levy MN (1996) Principles of physiology. Mosby-Year Book Inc, St. Louis, MO
Briggs MM, Schachat F (2000) Early specialization of the superfast myosin in extraocular and laryngeal muscles. J␣Exp Biol 203:2485–2494
Brown EJ, Schreiber SL (1996) A signaling pathway to translational control. Cell 86:517–520
Buckingham ME, Cohen A, Gros F (1976) Cytoplasmic distribution of pulse-labelled poly(A)-containing RNA, particularly 26 S RNA, during myoblast growth and differentiation. J Mol Biol 103:611–626
Buttrick PM, Kaplan ML, Kitsis RN, Leinwand LA (1993) Distinct behavior of cardiac myosin heavy chain gene constructs in vivo. Discordance with in vitro results. Circ Res 72:1211–1217
Campbell WG, Gordon SE, Carlson CJ, Pattison JS, Hamilton MT, Booth FW (2001) Differential global gene expression in red and white skeletal muscle. Am J Physiol Cell Physiol 280:C763–C768
Coirault C, Lambert F, Marchand-Adam S, Attal P, Chemla D, Lecarpentier Y (1999) Myosin molecular motor dysfunction in dystrophic mouse diaphragm. Am J Physiol 277:C1170–C1176
D’albis A, Couteaux R, Janmot C, Roulet A, Mira JC (1988) Regeneration after cardiotoxin injury of innervated and denervated slow and fast muscles of mammals. Myosin isoform analysis. Eur J Biochem 174:103–110
D’Albis A, Couteaux R, Janmot C, Mira JC (1989) Myosin isoform transitions in regeneration of fast and slow muscles during postnatal development of the rat. Dev Biol 135:320–325
Da Costa N, Beuzen N, Johnston I, McGillivray C, Sun YM, Chang KC (2000) The 5’-end of the porcine perinatal myosin heavy chain gene shows alternative splicing and is clustered with repeat elements. J Muscle Res Cell Motil 21:183–188
Diagana TT, North DL, Jabet C, Fiszman MY, Takeda S, Whalen RG (1997) The transcriptional activity of a muscle-specific promoter depends critically on the structure of the TATA element and its binding protein. J Mol Biol 265:480–493
The Fantom Consortium (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563
Gulick J, Subramaniam A, Neumann J, Robbins J (1991) Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem 266:9180–9185
Hashimoto S, Suzuki Y, Kasai Y, Morohoshi K, Yamada T, Sese J, Morishita S, Sugano S, Matsushima K (2004) 5’-end SAGE for the analysis of transcriptional start sites. Nat Biotechnol 22:1146–1149
Itoh K, Adelstein RS (1995) Neuronal cell expression of inserted isoforms of vertebrate nonmuscle myosin heavy chain II-B. J Biol Chem 270:14533–14540
Iyer V, Struhl K (1995) Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol 15:7059–7066
Lu BD, Allen DL, Leinwand LA, Lyons GE (1999) Spatial and temporal changes in myosin heavy chain gene expression in skeletal muscle development. Dev Biol 216:312–326
Lucas CA, Hoh JF (2003) Distribution of developmental myosin heavy chains in adult rabbit extraocular muscle: identification of a novel embryonic isoform absent in fetal limb. Invest Ophthalmol Vis Sci 44:2450–2456
Lyons GE, Ontell M, Cox R, Sassoon D, Buckingham M (1990a) The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol 111:1465–1476
Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M (1990b) Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol 111:2427–2436
Meric F, Hunt KK (2002) Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther 1:971–979
Morkin E (2000) Control of cardiac myosin heavy chain gene expression. Microsc Res Tech 50:522–531
Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N (2001) Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet 27:195–200
Nagai R, Kuro-o M, Babij P, Periasamy M (1989) Identification of two types of smooth muscle myosin heavy chain isoforms by cDNA cloning and immunoblot analysis. J Biol Chem 264:9734–9737
Oliva D, Venturella S, Passantino R, Feo S, Giallongo A (1995) Conserved alternative splicing in the 5’-untranslated region of the muscle-specific enolase gene. Primary structure of mRNAs, expression and influence of secondary structure on the translation efficiency. Eur J Biochem 232:141–149
Ontell M, Ontell MP, Sopper MM, Mallonga R, Lyons G, Buckingham M (1993) Contractile protein gene expression in primary myotybes of embyryonic hindlimb muscles. Development 117:1435–1444
Parker MH, Seale P, Rudnicki MA (2003) Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet 4:497–507
Periasamy M, Strehler EE, Garfinkel LI, Gubits RM, Ruiz-Opazo N, Nadal-Ginard B (1984) Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem 259:13595–13604
Pette D, Staron RS (2001) Transitions of muscle fiber phenotypic profiles. Histochem Cell Biol 115:359–372
Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Andrade FH (2001) Extraocular muscle is defined by a fundamentally distinct gene expression profile. Proc Natl Acad Sci USA 98:12062– 12067
Porter JD, Merriam AP, Leahy P, Gong B, Feuerman J, Cheng G, Khanna S (2004) Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle group-specific mechanisms in the pathogenesis of muscular dystrophy. Hum Mol Genet 13:257–269
Reggiani C, Brocks L, Wirtz P, Loermans H, te Kronnie G (1992) Myosin isoforms in hindlimb muscles of normal and dystrophic (ReJ129 dy/dy) mice. Muscle Nerve 15:199–208
Rindt H, Gulick J, Knotts S, Neumann J, Robbins J (1993) In␣vivo analysis of the murine β-myosin heavy chain gene promoter. J Biol Chem 268:5332–5338
Rindt H, Subramaniam A, Robbins J (1995) An in vivo analysis of transcriptional elements in the mouse α-myosin heavy chain gene promoter. Transgenic Res 4:397–405
Robert B, Daubas P, Akimenko MA, Cohen A, Garner I, Guenet JL, Buckingham M (1984) A single locus in the mouse encodes both myosin light chains 1 and 3, a second locus corresponds to a related pseudogene. Cell 39:129–140
Roy A, Exinger F, Losson R (1990) Cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol 10:5257–5270
Sartore S, Gorza L, Schiaffino S (1982) Fetal myosin heavy chains in regenerating muscle. Nature 298:294–296
Sartorius CA, Lu BD, Acakpo-Satchivi L, Jacobsen RP, Byrnes WC, Leinwand LA (1998) Myosin heavy chains IIa and IId are functionally distinct in the mouse. J Cell Biol 141:943–953
Standiford DM, Sun WT, Davis MB, Emerson CP Jr (2001) Positive and negative intronic regulatory elements control muscle-specific alternative exon splicing of Drosophila myosin heavy chain transcripts. Genetics 157:259–271
Subramaniam A, Gulick J, Neumann J, Knotts S, Robbins J␣(1993) Transgenic analysis of the thyroid-responsive elements in the α-cardiac myosin heavy chain gene promoter. J␣Biol Chem 268:4331–4336
Sun YM, da Costa N, Birrell R, Archibald AL, Alzuherri H, Chang KC (2001) Molecular and quantitative characterisation of the porcine embryonic myosin heavy chain gene. J␣Muscle Res Cell Motil 22:317–327
Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, Suyama A, Sakaki Y, Morishita S, Okubo K, Sugano S (2001) Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res 11:677–684
Swank DM, Bartoo ML, Knowles AF, Iliffe C, Bernstein SI, Molloy JE, Sparrow JC (2001) Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J Biol Chem 276:15117–15124
Takahashi M, Kawamoto S, Adelstein RS (1992) Evidence for inserted sequences in the head region of nonmuscle myosin specific to the nervous system. Cloning of the cDNA encoding the myosin heavy chain-B isoform of vertebrate nonmuscle myosin. J Biol Chem 267:17864–17871
Takeda S, North DL, Lakich MM, Russell SD, Whalen RG (1992) A possible regulatory role for conserved promoter motifs in an adult-specific muscle myosin gene from mouse. J Biol Chem 267:16957–16967
Takeda S, North DL, Diagana T, Miyagoe Y, Lakich MM, Whalen RG (1995) Myogenic regulatory factors can activate TATA-containing promoter elements via an E-box independent mechanism. J Biol Chem 270:15664–15670
The Mammalian Gene Collection Program (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99:16899–16903
Vikstrom KL, Factor SM, Leinwand LA (1996) Mice expressing mutant myosin heavy chains are a model for familial hypertrophic cardiomyopathy. Mol Med 2:556–567
Walro JM, Kucera J (1999) Why adult mammalian intrafusal and extrafusal fibers contain different myosin heavy-chain isoforms. Trends Neurosci 22:180–184
Warrick HM, Spudich JA (1987) Myosin structure and function in cell motility. Annu Rev Cell Biol 3:379–421
Weiss A, McDonough D, Wertman B, Acakpo-Satchivi L, Montgomery K, Kucherlapati R, Leinwand L, Krauter K (1999) Organization of human and mouse skeletal myosin heavy chain gene clusters is highly conserved. Proc Natl Acad Sci USA 96:2958–2963
Weydert A, Daubas P, Lazaridis I, Barton P, Garner I, Leader DP, Bonhomme F, Catalan J, Simon D, Guenet JL et al (1985) Genes for skeletal muscle myosin heavy chains are clustered and are not located on the same mouse chromosome as a cardiac myosin heavy chain gene. Proc Natl Acad Sci USA 82:7183–7187
Weydert A, Barton P, Harris AJ, Pinset C, Buckingham M (1987) Developmental pattern of mouse skeletal myosin heavy chain gene transcripts in vivo and in vitro. Cell 49:121–129
White SL, Zhou MY, Low RB, Periasamy M (1998) Myosin heavy chain isoform expression in rat smooth muscle development. Am J Physiol (Cell Physiol 44) 275:C581–C589
Yamada A, Yoshio M, Oiwa K, Nyitray L (2000) Catchin, a novel protein in molluscan catch muscles, is produced by alternative splicing from the myosin heavy chain gene. J Mol Biol 295:169–178
Zhang S, Bernstein SI (2001) Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev 101:35–45
Acknowledgments
This work was supported by Public Health Service grants NIH GM029090, NIH HL050560, and NIH AR048817 to L. A. L., and NIH 5 P60 DA011015, and NIH 5 R01 DA012845 to K. S. K. Training grant support to B. K. D. was from NIH HL007851. We thank Angelika Paul for cardiotoxin-damaged muscle RNA and mouse anatomy lessons. We thank Matthew Bell for 15.5 and 17.5 dpc mice; Allison Cleary and Doris Beylkin for neonates; Stephen Luckey, Brian Stauffer, Angelika Paul and John Konhilas for general assistance; and Dave Allen for tissue culture lessons. Joseph Heimiller helped with sequence analysis, Eric Robbins helped with DNA preparation and Lindsay Smith helped with PCR.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Dennehey, B.K., Leinwand, L.A. & Krauter, K.S. Diversity in transcriptional start site selection and alternative splicing affects the 5′-UTR of mouse striated muscle myosin transcripts. J Muscle Res Cell Motil 27, 559–575 (2006). https://doi.org/10.1007/s10974-006-9071-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10974-006-9071-8