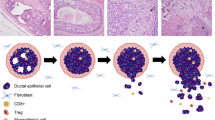
Abstract
Ductal carcinoma in situ (DCIS) is a non-invasive proliferative growth in the breast that serves as a non-obligate precursor to invasive ductal carcinoma. The widespread adoption of screening mammography has led to a steep increase in the detection of DCIS, which now comprises approximately 20% of new breast cancer diagnoses in the United States. Interestingly, the intratumoral heterogeneity (ITH) that has been observed in invasive breast cancers may have been established early in tumorigenesis, given the vast and varied ITH that has been detected in DCIS. This review will discuss the intratumoral heterogeneity of DCIS, focusing on the phenotypic and genomic heterogeneity of tumor cells, as well as the compositional heterogeneity of the tumor microenvironment. In addition, we will assess the spatial heterogeneity that is now being appreciated in these lesions, and summarize new approaches to evaluate heterogeneity of tumor and stromal cells in the context of their spatial organization. Importantly, we will discuss how a growing understanding of ITH has led to a more holistic appreciation of the complex biology of DCIS, specifically its evolution and natural history. Finally, we will consider ways in which our knowledge of DCIS ITH might be translated in the future to guide clinical care for DCIS patients.
Similar content being viewed by others
References
Wellings SR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst. 1973;50(5):1111–8.
Lee S, Mohsin SK, Mao S, Hilsenbeck SG, Medina D, Allred DC. Hormones, receptors, and growth in hyperplastic enlarged lobular units: early potential precursors of breast cancer. Breast Cancer Res. 2006;8(1):R6. https://doi.org/10.1186/bcr1367.
Tavassoli FA. Ductal carcinoma in situ: introduction of the concept of ductal intraepithelial neoplasia. Mod Pathol. 1998;11(2):140–54.
Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7(9):659–72.
Ellis IO. Intraductal proliferative lesions of the breast: morphology, associated risk and molecular biology. Mod Pathol. 2010;23(Suppl 2):S1–7. https://doi.org/10.1038/modpathol.2010.56.
Lakhani SR, Ellis. I.O., Schnitt, S.J., Tan, P.H., van de Vijver, M.J. WHO Classification of Tumours of the Breast. Lyon: IARC Press2012.
Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 2010;12(4):207. https://doi.org/10.1186/bcr2607.
Cowell CF, Weigelt B, Sakr RA, Ng CKY, Hicks J, King TA, et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol. 2013;7(5):859–69. https://doi.org/10.1016/j.molonc.2013.07.005.
Allred DC. Ductal carcinoma in situ: terminology, classification, and natural history. J Natl Cancer Inst Monogr. 2010;2010(41):134–8. https://doi.org/10.1093/jncimonographs/lgq035.
Bombonati A, Sgroi DC. The Molecular Pathology of Breast Cancer Progression. The Journal of pathology. 2011;223(2):307–17. https://doi.org/10.1002/path.2808.
NCCN. Breast Cancer Clinical Guidelines 2018: National Comprehensive Cancer Network2018.
American Cancer Society I. Breast Cancer Facts & Figures 2017-2018. Atlanta2017.
Allegra CJ, Aberle DR, Ganschow P, Hahn SM, Lee CN, Millon-Underwood S, et al. National Institutes of Health State-of-the-Science Conference statement: Diagnosis and Management of Ductal Carcinoma In Situ September 22-24, 2009. J Natl Cancer Inst. 2010;102(3):161–9. https://doi.org/10.1093/jnci/djp485.
Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, Decker T, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J Pathol. 1999;187(4):396–402. https://doi.org/10.1002/(sici)1096-9896(199903)187:4<396::aid-path286>3.0.co;2-l.
Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100(10):5974–9. https://doi.org/10.1073/pnas.0931261100.
Vincent-Salomon A, Lucchesi C, Gruel N, Raynal V, Pierron G, Goudefroye R, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(7):1956–65. https://doi.org/10.1158/1078-0432.ccr-07-1465.
Committee TCC. Consensus conference on the classification of ductal carcinoma in situ. Human pathology. 1997;28(11):1221–5.
Leong AS, Sormunen RT, Vinyuvat S, Hamdani RW, Suthipintawong C. Biologic markers in ductal carcinoma in situ and concurrent infiltrating carcinoma. A comparison of eight contemporary grading systems. Am J Clin Pathol. 2001;115(5):709–18. https://doi.org/10.1309/pj7h-a52v-m3xb-v94y.
Leonard GD, Swain SM. Ductal carcinoma in situ, complexities and challenges. J Natl Cancer Inst. 2004;96(12):906–20.
Pinder SE. Ductal carcinoma in situ (DCIS): pathological features, differential diagnosis, prognostic factors and specimen evaluation. Mod Pathol. 2010;23(Suppl 2):S8–13. https://doi.org/10.1038/modpathol.2010.40.
Pinder SE, Duggan C, Ellis IO, Cuzick J, Forbes JF, Bishop H, et al. A new pathological system for grading DCIS with improved prediction of local recurrence: results from the UKCCCR/ANZ DCIS trial. British journal of cancer. 2010;103(1):94–100. https://doi.org/10.1038/sj.bjc.6605718.
Brown JP, Pinder SE. Ductal carcinoma in situ: current morphological and molecular subtypes. Diagnostic Histopathology. 2012;18(3):112–8. https://doi.org/10.1016/j.mpdhp.2012.01.001.
Makki J. Diversity of Breast Carcinoma: Histological Subtypes and Clinical Relevance. Clinical Medicine Insights Pathology. 2015;8:23–31. https://doi.org/10.4137/CPath.S31563.
Gorringe KL, Fox SB. Ductal Carcinoma In Situ Biology, Biomarkers, and Diagnosis. Frontiers in Oncology. 2017;7:248. https://doi.org/10.3389/fonc.2017.00248.
Lennington WJ, Jensen RA, Dalton LW, Page DL. Ductal carcinoma in situ of the breast. Heterogeneity of individual lesions. Cancer. 1994;73(1):118–24.
Quinn CM, Ostrowski JL. Cytological and architectural heterogeneity in ductal carcinoma in situ of the breast. Journal of Clinical Pathology. 1997;50(7):596–9.
Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee S, Perou CM, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(2):370–8. https://doi.org/10.1158/1078-0432.CCR-07-1127.
Perez AA, Balabram D, Salles Mde A, Gobbi H. Ductal carcinoma in situ of the breast: correlation between histopathological features and age of patients. Diagnostic pathology. 2014;9:227. https://doi.org/10.1186/s13000-014-0227-3.
Scripcaru G, Zardawi IM. Mammary Ductal Carcinoma In Situ: A Fresh Look at Architectural Patterns. International Journal of Surgical Oncology. 2012;2012:979521. https://doi.org/10.1155/2012/979521.
Bane A. Ductal Carcinoma In Situ: What the Pathologist Needs to Know and Why. International Journal of Breast Cancer. 2013;2013:914053. https://doi.org/10.1155/2013/914053.
Chapman J-AW, Miller NA, Lickley HLA, Qian J, Christens-Barry WA, Fu Y et al. Ductal carcinoma in situ of the breast (DCIS) with heterogeneity of nuclear grade: prognostic effects of quantitative nuclear assessment. BMC Cancer. 2007;7:174-. doi:10.1186/1471-2407-7-174.
Pape-Zambito D, Jiang Z, Wu H, Devarajan K, Slater CM, Cai KQ, et al. Identifying a highly-aggressive DCIS subgroup by studying intra-individual DCIS heterogeneity among invasive breast cancer patients. PloS one. 2014;9(6):e100488. https://doi.org/10.1371/journal.pone.0100488.
Lari SA, Kuerer HM. Biological Markers in DCIS and Risk of Breast Recurrence: A Systematic Review. Journal of Cancer. 2011;2:232–61.
McCarty KS Jr, Miller LS, Cox EB, Konrath J, McCarty KS Sr. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Archives of pathology & laboratory medicine. 1985;109(8):716–21.
Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS). Pathol Res Pract. 1993;189(8):862–6. https://doi.org/10.1016/S0344-0338(11)81095-2.
Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11(2):155–68.
Smith KL, Robbins PD, Dawkins HJ, Papadimitriou JM, Redmond SL, Carrello S, et al. c-erbB-2 amplification in breast cancer: detection in formalin-fixed, paraffin-embedded tissue by in situ hybridization. Human pathology. 1994;25(4):413–8.
Latta EK, Tjan S, Parkes RK, O'Malley FP. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Mod Pathol. 2002;15(12):1318–25. https://doi.org/10.1097/01.mp.0000038462.62634.b1.
Park K, Han S, Kim HJ, Kim J, Shin E. HER2 status in pure ductal carcinoma in situ and in the intraductal and invasive components of invasive ductal carcinoma determined by fluorescence in situ hybridization and immunohistochemistry. Histopathology. 2006;48(6):702–7. https://doi.org/10.1111/j.1365-2559.2006.02403.x.
Van Bockstal M, Lambein K, Denys H, Braems G, Nuyts A, Van den Broecke R, et al. Histopathological characterization of ductal carcinoma in situ (DCIS) of the breast according to HER2 amplification status and molecular subtype. Virchows Archiv : an international journal of pathology. 2014;465(3):275–89. https://doi.org/10.1007/s00428-014-1609-3.
Ringberg A, Anagnostaki L, Anderson H, Idvall I, Ferno M. South Sweden Breast Cancer G. Cell biological factors in ductal carcinoma in situ (DCIS) of the breast-relationship to ipsilateral local recurrence and histopathological characteristics. Eur J Cancer. 2001;37(12):1514–22.
Lebeau A, Unholzer A, Amann G, Kronawitter M, Bauerfeind I, Sendelhofert A, et al. EGFR, HER-2/neu, cyclin D1, p21 and p53 in correlation to cell proliferation and steroid hormone receptor status in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2003;79(2):187–98.
Wilson GR, Cramer A, Welman A, Knox F, Swindell R, Kawakatsu H, et al. Activated c-SRC in ductal carcinoma in situ correlates with high tumour grade, high proliferation and HER2 positivity. British journal of cancer. 2006;95(10):1410–4. https://doi.org/10.1038/sj.bjc.6603444.
Kerlikowske K, Molinaro AM, Gauthier ML, Berman HK, Waldman F, Bennington J, et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. J Natl Cancer Inst. 2010;102(9):627–37. https://doi.org/10.1093/jnci/djq101.
Rakovitch E, Nofech-Mozes S, Hanna W, Narod S, Thiruchelvam D, Saskin R, et al. HER2/neu and Ki-67 expression predict non-invasive recurrence following breast-conserving therapy for ductal carcinoma in situ. British journal of cancer. 2012;106(6):1160–5. https://doi.org/10.1038/bjc.2012.41.
Buckley N, Boyle D, McArt D, Irwin G, Harkin DP, Lioe T et al. Molecular classification of non-invasive breast lesions for personalised therapy and chemoprevention. Oncotarget. 2015;6(41):43244-54. doi:10.18632/oncotarget.6525.
Williams KE, Barnes NL, Cramer A, Johnson R, Cheema K, Morris J, et al. Molecular phenotypes of DCIS predict overall and invasive recurrence. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26(5):1019–25. https://doi.org/10.1093/annonc/mdv062.
Poulakaki N, Makris GM, Papanota AM, Marineli F, Marinelis A, Battista MJ et al. Ki-67 Expression as a Factor Predicting Recurrence of Ductal Carcinoma In Situ of the Breast: A Systematic Review and Meta-Analysis. Clinical breast cancer. 2018;18(2):157-67.e6. doi:10.1016/j.clbc.2017.12.007.
Done SJ, Arneson NC, Ozcelik H, Redston M, Andrulis IL. p53 mutations in mammary ductal carcinoma in situ but not in epithelial hyperplasias. Cancer Res. 1998;58(4):785–9.
Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih Ie M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24(9):1248–53. https://doi.org/10.1038/modpathol.2011.85.
Pang JB, Savas P, Fellowes AP, Mir Arnau G, Kader T, Vedururu R, et al. Breast ductal carcinoma in situ carry mutational driver events representative of invasive breast cancer. Mod Pathol. 2017;30(7):952–63. https://doi.org/10.1038/modpathol.2017.21.
Rajan PB, Scott DJ, Perry RH, Griffith CD. p53 protein expression in ductal carcinoma in situ (DCIS) of the breast. Breast Cancer Res Treat. 1997;42(3):283–90.
Lukas J, Niu N, Press MF. p53 mutations and expression in breast carcinoma in situ. Am J Pathol. 2000;156(1):183–91. https://doi.org/10.1016/S0002-9440(10)64718-9.
Poller DN, Roberts EC, Bell JA, Elston CW, Blamey RW, Ellis IO. p53 protein expression in mammary ductal carcinoma in situ: relationship to immunohistochemical expression of estrogen receptor and c-erbB-2 protein. Human pathology. 1993;24(5):463–8.
Marchetti A, Buttitta F, Pellegrini S, Campani D, Cecchetti D, Bistocchi M. P53 and C-erbb-2 alterations in in-situ and invasive ductal breast carcinomas - a genetic and immunohistochemical analysis. International journal of oncology. 1995;7(2):343–7.
Meijnen P, Peterse JL, Antonini N, Rutgers EJT, van de Vijver MJ. Immunohistochemical categorisation of ductal carcinoma in situ of the breast. British journal of cancer. 2008;98(1):137–42. https://doi.org/10.1038/sj.bjc.6604112.
Gerdes MJ, Gokmen-Polar Y, Sui Y, Pang AS, LaPlante N, Harris AL, et al. Single-cell heterogeneity in ductal carcinoma in situ of breast. Mod Pathol. 2018;31(3):406–17. https://doi.org/10.1038/modpathol.2017.143.
Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, et al. Promoter CpG island hypermethylation during breast cancer progression. Virchows Archiv : an international journal of pathology. 2011;458(1):73–84. https://doi.org/10.1007/s00428-010-1013-6.
Johnson KC, Koestler DC, Fleischer T, Chen P, Jenson EG, Marotti JD, et al. DNA methylation in ductal carcinoma in situ related with future development of invasive breast cancer. Clin Epigenetics. 2015;7:75. https://doi.org/10.1186/s13148-015-0094-0.
Yoon HJ, Kim Y, Kim BS. Intratumoral metabolic heterogeneity predicts invasive components in breast ductal carcinoma in situ. Eur Radiol. 2015;25(12):3648–58. https://doi.org/10.1007/s00330-015-3761-9.
Kaur H, Mao S, Li Q, Sameni M, Krawetz SA, Sloane BF, et al. RNA-Seq of human breast ductal carcinoma in situ models reveals aldehyde dehydrogenase isoform 5A1 as a novel potential target. PloS one. 2012;7(12):e50249. https://doi.org/10.1371/journal.pone.0050249.
Eswaran J, Horvath A, Godbole S, Reddy SD, Mudvari P, Ohshiro K, et al. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep. 2013;3:1689. https://doi.org/10.1038/srep01689.
Abba MC, Gong T, Lu Y, Lee J, Zhong Y, Lacunza E, et al. A Molecular Portrait of High-Grade Ductal Carcinoma In Situ. Cancer Res. 2015;75(18):3980–90. https://doi.org/10.1158/0008-5472.CAN-15-0506.
Elsarraj HS, Hong Y, Valdez KE, Michaels W, Hook M, Smith WP, et al. Expression profiling of in vivo ductal carcinoma in situ progression models identified B cell lymphoma-9 as a molecular driver of breast cancer invasion. Breast Cancer Res. 2015;17:128. https://doi.org/10.1186/s13058-015-0630-z.
Locke WJ, Clark SJ. Epigenome remodelling in breast cancer: insights from an early in vitro model of carcinogenesis. Breast Cancer Res. 2012;14(6):215. https://doi.org/10.1186/bcr3237.
Moelans CB, de Groot JS, Pan X, van der Wall E, van Diest PJ. Clonal intratumor heterogeneity of promoter hypermethylation in breast cancer by MS-MLPA. Mod Pathol. 2014;27(6):869–74. https://doi.org/10.1038/modpathol.2013.207.
Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342(6163):1243259. https://doi.org/10.1126/science.1243259.
Fessenden M. Metabolomics: Small molecules, single cells. Nature. 2016;540(7631):153–5. https://doi.org/10.1038/540153a.
Emara S, Amer S, Ali A, Abouleila Y, Oga A, Masujima T. Single-Cell Metabolomics. Adv Exp Med Biol. 2017;965:323–43. https://doi.org/10.1007/978-3-319-47656-8_13.
Almuhaideb A, Papathanasiou N, Bomanji J. (18)F-FDG PET/CT Imaging In Oncology. Annals of Saudi Medicine. 2011;31(1):3–13. https://doi.org/10.4103/0256-4947.75771.
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. https://doi.org/10.1038/nmeth.1315.
Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13(4):599–604. https://doi.org/10.1038/nprot.2017.149.
Nielsen KV, Blichert-Toft M, Andersen J. Chromosome analysis of in situ breast cancer. Acta oncologica (Stockholm, Sweden). 1989;28(6):919-22.
Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nature genetics. 2004;36(9):984–8. https://doi.org/10.1038/ng1409.
Afghahi A, Forgo E, Mitani AA, Desai M, Varma S, Seto T, et al. Chromosomal copy number alterations for associations of ductal carcinoma in situ with invasive breast cancer. Breast Cancer Res. 2015;17:108. https://doi.org/10.1186/s13058-015-0623-y.
Visscher DW, Wallis TL, Crissman JD. Evaluation of chromosome aneuploidy in tissue sections of preinvasive breast carcinomas using interphase cytogenetics. Cancer. 1996;77(2):315–20. https://doi.org/10.1002/(sici)1097-0142(19960115)77:2<315::aid-cncr14>3.0.co;2-4.
Murphy DS, Hoare SF, Going JJ, Mallon EE, George WD, Kaye SB, et al. Characterization of extensive genetic alterations in ductal carcinoma in situ by fluorescence in situ hybridization and molecular analysis. J Natl Cancer Inst. 1995;87(22):1694–704.
Meeker AK, Hicks JL, Iacobuzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, et al. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10(10):3317–26. https://doi.org/10.1158/1078-0432.ccr-0984-03.
Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, et al. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. Am J Pathol. 2012;181(5):1807–22. https://doi.org/10.1016/j.ajpath.2012.07.012.
Jang M, Kim E, Choi Y, Lee H, Kim Y, Kim J, et al. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14(4):R115. https://doi.org/10.1186/bcr3239.
Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–21.
James LA, Mitchell EL, Menasce L, Varley JM. Comparative genomic hybridisation of ductal carcinoma in situ of the breast: identification of regions of DNA amplification and deletion in common with invasive breast carcinoma. Oncogene. 1997;14(9):1059–65. https://doi.org/10.1038/sj.onc.1200923.
Iakovlev VV, Arneson NC, Wong V, Wang C, Leung S, Iakovleva G, et al. Genomic differences between pure ductal carcinoma in situ of the breast and that associated with invasive disease: a calibrated aCGH study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(14):4446–54. https://doi.org/10.1158/1078-0432.ccr-07-4960.
Stratton MR, Collins N, Lakhani SR, Sloane JP. Loss of heterozygosity in ductal carcinoma in situ of the breast. J Pathol. 1995;175(2):195–201. https://doi.org/10.1002/path.1711750207.
Pan A, Zhou Y, Mu K, Liu Y, Sun F, Li P, et al. Detection of gene copy number alterations in DCIS and invasive breast cancer by QM-FISH. Am J Transl Res. 2016;8(11):4994–5004.
Fischer A, Vazquez-Garcia I, Illingworth CJR, Mustonen V. High-definition reconstruction of clonal composition in cancer. Cell Rep. 2014;7(5):1740–52. https://doi.org/10.1016/j.celrep.2014.04.055.
Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10(8):e1003665. https://doi.org/10.1371/journal.pcbi.1003665.
Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–8. https://doi.org/10.1038/nmeth.2883.
Schwarz RF, Trinh A, Sipos B, Brenton JD, Goldman N, Markowetz F. Phylogenetic quantification of intra-tumour heterogeneity. PLoS Comput Biol. 2014;10(4):e1003535. https://doi.org/10.1371/journal.pcbi.1003535.
Gupta RG, Somer RA. Intratumor Heterogeneity: Novel Approaches for Resolving Genomic Architecture and Clonal Evolution. Mol Cancer Res. 2017;15(9):1127–37. https://doi.org/10.1158/1541-7786.MCR-17-0070.
Ortega MA, Poirion O, Zhu X, Huang S, Wolfgruber TK, Sebra R, et al. Using single-cell multiple omics approaches to resolve tumor heterogeneity. Clinical and translational medicine. 2017;6(1):46. https://doi.org/10.1186/s40169-017-0177-y.
Schwartz R, Schaffer AA. The evolution of tumour phylogenetics: principles and practice. Nat Rev Genet. 2017;18(4):213–29. https://doi.org/10.1038/nrg.2016.170.
Niida A, Nagayama S, Miyano S, Mimori K. Understanding intratumor heterogeneity by combining genome analysis and mathematical modeling. Cancer Sci. 2018;109(4):884–92. https://doi.org/10.1111/cas.13510.
Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–92. https://doi.org/10.1056/NEJMoa1113205.
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014;46(3):225–33. https://doi.org/10.1038/ng.2891.
Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–9. https://doi.org/10.1126/science.1256930.
Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–9. https://doi.org/10.1038/nm.3886.
El-Kebir M, Satas G, Oesper L, Raphael BJ. Inferring the Mutational History of a Tumor Using Multi-state Perfect Phylogeny Mixtures. Cell Syst. 2016;3(1):43–53. https://doi.org/10.1016/j.cels.2016.07.004.
Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nature communications. 2017;8(1):268. https://doi.org/10.1038/s41467-017-00296-y.
Foschini MP, Morandi L, Leonardi E, Flamminio F, Ishikawa Y, Masetti R, et al. Genetic clonal mapping of in situ and invasive ductal carcinoma indicates the field cancerization phenomenon in the breast. Human pathology. 2013;44(7):1310–9. https://doi.org/10.1016/j.humpath.2012.09.022.
Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, et al. Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing. Cell. 2018;172(1-2):205–17 e12. https://doi.org/10.1016/j.cell.2017.12.007.
Shannon CE. A mathematical theory of communication. The Bell System Technical Journal. 1948;27(3):379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x.
Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–8.
Greaves M. Nothing in cancer makes sense except. BMC Biol. 2018;16(1):22. https://doi.org/10.1186/s12915-018-0493-8.
Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA. Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces. Cancer Res. 2016;76(11):3136–44. https://doi.org/10.1158/0008-5472.can-15-2962.
Sameni M, Cavallo-Medved D, Franco OE, Chalasani A, Ji K, Aggarwal N, et al. Pathomimetic avatars reveal divergent roles of microenvironment in invasive transition of ductal carcinoma in situ. Breast Cancer Res. 2017;19(1):56. https://doi.org/10.1186/s13058-017-0847-0.
Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13(6):227. https://doi.org/10.1186/bcr2912.
Pienta KJ, McGregor N, Axelrod R, Axelrod DE. Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Translational oncology. 2008;1(4):158–64.
Kim IS, Zhang XH. One microenvironment does not fit all: heterogeneity beyond cancer cells. Cancer Metastasis Rev. 2016;35(4):601–29. https://doi.org/10.1007/s10555-016-9643-z.
Hui L, Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015;368(1):7–13. https://doi.org/10.1016/j.canlet.2015.07.039.
Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8(5):761–73. https://doi.org/10.7150/jca.17648.
Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. Journal of cell science. 2012;125(Pt 23):5591–6. https://doi.org/10.1242/jcs.116392.
Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC medicine. 2015;13:45. https://doi.org/10.1186/s12916-015-0278-7.
McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nature cell biology. 2014;16(8):717–27. https://doi.org/10.1038/ncb3015.
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine. 2013;19(11):1423–37. https://doi.org/10.1038/nm.3394.
Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends in genetics : TIG. 2009;25(1):30–8. https://doi.org/10.1016/j.tig.2008.10.012.
Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514(7520):54–8. https://doi.org/10.1038/nature13556.
Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5(8):806–20. https://doi.org/10.1158/2159-8290.cd-15-0439.
Takahashi K, Ehata S, Koinuma D, Morishita Y, Soda M, Mano H, et al. Pancreatic tumor microenvironment confers highly malignant properties on pancreatic cancer cells. Oncogene. 2018; https://doi.org/10.1038/s41388-018-0144-0.
Naeem S, Duffy JE, Zavaleta E. The functions of biological diversity in an age of extinction. Science. 2012;336(6087):1401–6. https://doi.org/10.1126/science.1215855.
Ives AR, Carpenter SR. Stability and diversity of ecosystems. Science. 2007;317(5834):58–62. https://doi.org/10.1126/science.1133258.
Harrison PA, Berry PM, Simpson G, Haslett JR, Blicharska M, Bucur M, et al. Linkages between biodiversity attributes and ecosystem services: A systematic review. Ecosystem Services. 2014;9:191–203. https://doi.org/10.1016/j.ecoser.2014.05.006.
Tilman D, Isbell F, Cowles JM. Biodiversity and Ecosystem Functioning. Annual Review of Ecology, Evolution, and Systematics. 2014;45(1):471–93. https://doi.org/10.1146/annurev-ecolsys-120213-091917.
de Vos MGJ, Zagorski M, McNally A, Bollenbach T. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc Natl Acad Sci U S A. 2017;114(40):10666–71. https://doi.org/10.1073/pnas.1713372114.
Calcagno V, Jarne P, Loreau M, Mouquet N, David P. Diversity spurs diversification in ecological communities. Nature communications. 2017;8:15810. https://doi.org/10.1038/ncomms15810.
Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. https://doi.org/10.1016/j.cell.2010.03.014.
Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–77. https://doi.org/10.1016/j.cell.2013.03.021.
Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, et al. A Big Bang model of human colorectal tumor growth. Nature genetics. 2015;47(3):209–16. https://doi.org/10.1038/ng.3214.
Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nature genetics. 2016;48(10):1119–30. https://doi.org/10.1038/ng.3641.
Jimenez-Sanchez A, Memon D, Pourpe S, Veeraraghavan H, Li Y, Vargas HA et al. Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell. 2017;170(5):927-38.e20. doi:10.1016/j.cell.2017.07.025.
Heindl A, Lan C, Rodrigues DN, Koelble K, Yuan Y. Similarity and diversity of the tumor microenvironment in multiple metastases: critical implications for overall and progression-free survival of high-grade serous ovarian cancer. Oncotarget. 2016;7(44):71123-35. doi:10.18632/oncotarget.12106.
Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–6. https://doi.org/10.1016/j.cell.2006.01.007.
Wells DK, Chuang Y, Knapp LM, Brockmann D, Kath WL, Leonard JN. Spatial and functional heterogeneities shape collective behavior of tumor-immune networks. PLoS Comput Biol. 2015;11(4):e1004181. https://doi.org/10.1371/journal.pcbi.1004181.
Natrajan R, Sailem H, Mardakheh FK, Arias Garcia M, Tape CJ, Dowsett M, et al. Microenvironmental Heterogeneity Parallels Breast Cancer Progression: A Histology–Genomic Integration Analysis. PLOS Medicine. 2016;13(2):e1001961. https://doi.org/10.1371/journal.pmed.1001961.
Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, Xavier JB. Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci U S A. 2017;114(11):2934–9. https://doi.org/10.1073/pnas.1700600114.
Campbell MJ, Baehner F, O'Meara T, Ojukwu E, Han B, Mukhtar R, et al. Characterizing the immune microenvironment in high-risk ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2017;161(1):17–28. https://doi.org/10.1007/s10549-016-4036-0.
Lyons YA, Wu SY, Overwijk WW, Baggerly KA, Sood AK. Immune cell profiling in cancer: molecular approaches to cell-specific identification. npj Precision Oncology. 2017;1(1):26. doi:10.1038/s41698-017-0031-0.
Behbod F, Xian W, Shaw CA, Hilsenbeck SG, Tsimelzon A, Rosen JM. Transcriptional profiling of mammary gland side population cells. Stem cells (Dayton, Ohio). 2006;24(4):1065-74. doi:10.1634/stemcells.2005-0375.
Kendrick H, Regan JL, Magnay FA, Grigoriadis A, Mitsopoulos C, Zvelebil M, et al. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics. 2008;9:591. https://doi.org/10.1186/1471-2164-9-591.
Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3(1):109–18. https://doi.org/10.1016/j.stem.2008.05.018.
Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12(2):R21. https://doi.org/10.1186/bcr2560.
Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14(5):R134. https://doi.org/10.1186/bcr3334.
dos Santos CO, Rebbeck C, Rozhkova E, Valentine A, Samuels A, Kadiri LR, et al. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc Natl Acad Sci U S A. 2013;110(18):7123–30. https://doi.org/10.1073/pnas.1303919110.
Russell TD, Jindal S, Agunbiade S, Gao D, Troxell M, Borges VF, et al. Myoepithelial cell differentiation markers in ductal carcinoma in situ progression. Am J Pathol. 2015;185(11):3076–89. https://doi.org/10.1016/j.ajpath.2015.07.004.
Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nature communications. 2017;8(1):1627. https://doi.org/10.1038/s41467-017-01560-x.
Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. https://doi.org/10.1016/j.ymeth.2014.08.016.
Carstens JL. Correa de Sampaio P, Yang D, Barua S, Wang H. Rao A et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nature communications. 2017;8:15095. https://doi.org/10.1038/ncomms15095.
Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget M-A, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Scientific Reports. 2017;7(1):13380. https://doi.org/10.1038/s41598-017-13942-8.
Chang Q, Ornatsky OI, Siddiqui I, Loboda A, Baranov VI, Hedley DW. Imaging Mass Cytometry. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2017;91(2):160–9. https://doi.org/10.1002/cyto.a.23053.
Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Meth. 2014;11(4):417–22. https://doi.org/10.1038/nmeth.2869.
Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–42. https://doi.org/10.1038/nm.3488.
Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Meth. 2017;advance online publication. doi:10.1038/nmeth.4391. http://www.nature.com/nmeth/journal/vaop/ncurrent/abs/nmeth.4391.html#supplementary-information.
Noguchi S, Motomura K, Inaji H, Imaoka S, Koyama H. Clonal analysis of human breast cancer by means of the polymerase chain reaction. Cancer Res. 1992;52(23):6594–7.
Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12(2):160–70. https://doi.org/10.1016/j.ccr.2007.06.013.
Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–22. https://doi.org/10.1038/nature09781.
Zhang M, Lee AV, Rosen JM. The Cellular Origin and Evolution of Breast Cancer. Cold Spring Harb Perspect Med. 2017;7(3). doi:10.1101/cshperspect.a027128.
Teixeira MR, Pandis N, Bardi G, Andersen JA, Mandahl N, Mitelman F, et al. Cytogenetic analysis of multifocal breast carcinomas: detection of karyotypically unrelated clones as well as clonal similarities between tumour foci. British journal of cancer. 1994;70(5):922–7.
Garcia SB, Novelli M, Wright NA. The clonal origin and clonal evolution of epithelial tumours. Int J Exp Pathol. 2000;81(2):89–116.
Shibata A, Tsai YC, Press MF, Henderson BE, Jones PA, Ross RK. Clonal analysis of bilateral breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 1996;2(4):743–8.
Trujillo KA, Hines WC, Vargas KM, Jones AC, Joste NE, Bisoffi M, et al. Breast field cancerization: isolation and comparison of telomerase-expressing cells in tumor and tumor adjacent, histologically normal breast tissue. Mol Cancer Res. 2011;9(9):1209–21. https://doi.org/10.1158/1541-7786.MCR-10-0424.
Polyak K. Breast cancer: origins and evolution. The Journal of clinical investigation. 2007;117(11):3155–63. https://doi.org/10.1172/jci33295.
Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–91. https://doi.org/10.1016/j.stem.2014.02.006.
Skibinski A, Kuperwasser C. The origin of breast tumor heterogeneity. Oncogene. 2015;34(42):5309–16. https://doi.org/10.1038/onc.2014.475.
Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12(2):133–43. https://doi.org/10.1038/nrc3184.
Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–13. https://doi.org/10.1038/nature10762.
Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–70. https://doi.org/10.1038/nature03482.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. https://doi.org/10.1073/pnas.0530291100.
Lampejo OT, Barnes DM, Smith P, Millis RR. Evaluation of infiltrating ductal carcinomas with a DCIS component: correlation of the histologic type of the in situ component with grade of the infiltrating component. Semin Diagn Pathol. 1994;11(3):215–22.
Fujii H, Marsh C, Cairns P, Sidransky D, Gabrielson E. Genetic divergence in the clonal evolution of breast cancer. Cancer Res. 1996;56(7):1493–7.
Gupta SK, Douglas-Jones AG, Fenn N, Morgan JM, Mansel RE. The clinical behavior of breast carcinoma is probably determined at the preinvasive stage (ductal carcinoma in situ). Cancer. 1997;80(9):1740–5.
Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1(5):362–75.
Reis-Filho JS, Lakhani SR. The diagnosis and management of pre-invasive breast disease: genetic alterations in pre-invasive lesions. Breast Cancer Res. 2003;5(6):313–9. https://doi.org/10.1186/bcr650.
Hwang ES, DeVries S, Chew KL, Moore DH 2nd, Kerlikowske K, Thor A, et al. Patterns of chromosomal alterations in breast ductal carcinoma in situ. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10(15):5160–7. https://doi.org/10.1158/1078-0432.CCR-04-0165.
Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66(8):4065–78. https://doi.org/10.1158/0008-5472.CAN-05-4083.
Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of In Situ to Invasive Breast Carcinoma Transition. Cancer cell. 2008;13(5):394–406. https://doi.org/10.1016/j.ccr.2008.03.007.
Gao Y, Niu Y, Wang X, Wei L, Lu S. Genetic changes at specific stages of breast cancer progression detected by comparative genomic hybridization. Journal of molecular medicine (Berlin, Germany). 2009;87(2):145-52. doi:10.1007/s00109-008-0408-1.
Moelans CB, de Weger RA, Monsuur HN, Maes AH, van Diest PJ. Molecular differences between ductal carcinoma in situ and adjacent invasive breast carcinoma: a multiplex ligation-dependent probe amplification study. Analytical cellular pathology (Amsterdam). 2010;33(3):165–73. https://doi.org/10.3233/acp-clo-2010-0546.
Johnson CE, Gorringe KL, Thompson ER, Opeskin K, Boyle SE, Wang Y, et al. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Res Treat. 2012;133(3):889–98. https://doi.org/10.1007/s10549-011-1835-1.
Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, et al. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72(17):4574–86. https://doi.org/10.1158/0008-5472.CAN-12-0636.
Liao S, Desouki MM, Gaile DP, Shepherd L, Nowak NJ, Conroy J, et al. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes Chromosomes Cancer. 2012;51(12):1067–78. https://doi.org/10.1002/gcc.21991.
Rohilla M, Bal A, Singh G, Joshi K. Prediction of heterogeneity in breast cancer immunophenotype at ductal carcinoma in situ stage? Journal of cancer research and therapeutics. 2016;12(4):1249–56. https://doi.org/10.4103/0973-1482.199541.
Espina V, Mariani BD, Gallagher RI, Tran K, Banks S, Wiedemann J, et al. Malignant Precursor Cells Pre-Exist in Human Breast DCIS and Require Autophagy for Survival. PloS one. 2010;5(4):e10240. https://doi.org/10.1371/journal.pone.0010240.
Kim SY, Jung SH, Kim MS, Baek IP, Lee SH, Kim TM et al. Genomic differences between pure ductal carcinoma in situ and synchronous ductal carcinoma in situ with invasive breast cancer. Oncotarget. 2015;6(10):7597-607. doi:10.18632/oncotarget.3162.
Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89(19):9064–8.
Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137(1):231–45.
Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32. https://doi.org/10.1016/j.ccr.2004.06.010.
Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: the role of extracellular matrix and its degrading enzymes. Cold Spring Harbor symposia on quantitative biology. 2005;70:343–56. https://doi.org/10.1101/sqb.2005.70.013.
Allen MD, Thomas GJ, Clark S, Dawoud MM, Vallath S, Payne SJ, et al. Altered microenvironment promotes progression of preinvasive breast cancer: myoepithelial expression of alphavbeta6 integrin in DCIS identifies high-risk patients and predicts recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(2):344–57. https://doi.org/10.1158/1078-0432.ccr-13-1504.
Gil Del Alcazar CR, Huh SJ, Ekram MB, Trinh A, Liu LL, Beca F, et al. Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition. Cancer Discov. 2017;7(10):1098–115. https://doi.org/10.1158/2159-8290.cd-17-0222.
Miroshnikova YA, Rozenberg GI, Cassereau L, Pickup M, Mouw JK, Ou G, et al. alpha5beta1-Integrin promotes tension-dependent mammary epithelial cell invasion by engaging the fibronectin synergy site. Molecular biology of the cell. 2017;28(22):2958–77. https://doi.org/10.1091/mbc.E17-02-0126.
Allott EH, Geradts J, Sun X, Cohen SM, Zirpoli GR, Khoury T, et al. Intratumoral heterogeneity as a source of discordance in breast cancer biomarker classification. Breast Cancer Research. 2016;18(1):68. https://doi.org/10.1186/s13058-016-0725-1.
Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson AN, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. Jama. 2015;313(11):1122–32. https://doi.org/10.1001/jama.2015.1405.
Elmore JG, Nelson HD, Pepe MS, et al. Variability in pathologists' interpretations of individual breast biopsy slides: A population perspective. Annals of Internal Medicine. 2016;164(10):649–55. https://doi.org/10.7326/M15-0964.
Leonard G. Discordant interpretations of breast biopsy specimens by pathologists. Jama. 2015;314(1):82–3. https://doi.org/10.1001/jama.2015.6224.
Dillon MF, McDermott EW, Quinn CM, O'Doherty A, O'Higgins N, Hill AD. Predictors of invasive disease in breast cancer when core biopsy demonstrates DCIS only. Journal of surgical oncology. 2006;93(7):559–63. https://doi.org/10.1002/jso.20445.
Brennan ME, Turner RM, Ciatto S, Marinovich ML, French JR, Macaskill P, et al. Ductal carcinoma in situ at core-needle biopsy: meta-analysis of underestimation and predictors of invasive breast cancer. Radiology. 2011;260(1):119–28. https://doi.org/10.1148/radiol.11102368.
Sim YT, Litherland J, Lindsay E, Hendry P, Brauer K, Dobson H, et al. Upgrade of ductal carcinoma in situ on core biopsies to invasive disease at final surgery: a retrospective review across the Scottish Breast Screening Programme. Clinical radiology. 2015;70(5):502–6. https://doi.org/10.1016/j.crad.2014.12.019.
Shi B, Grimm LJ, Mazurowski MA, Baker JA, Marks JR, King LM et al. Prediction of Occult Invasive Disease in Ductal Carcinoma in Situ Using Deep Learning Features. Journal of the American College of Radiology : JACR. 2018;15(3 Pt B):527-34. doi:10.1016/j.jacr.2017.11.036.
Seol H, Lee HJ, Choi Y, Lee HE, Kim YJ, Kim JH, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol. 2012;25(7):938–48. https://doi.org/10.1038/modpathol.2012.36.
Song H, Kim TO, Ma SY, Park JH, Choi JH, Kim JH, et al. Intratumoral heterogeneity impacts the response to anti-neu antibody therapy. BMC Cancer. 2014;14:647. https://doi.org/10.1186/1471-2407-14-647.
Ng CK, Martelotto LG, Gauthier A, Wen HC, Piscuoglio S, Lim RS, et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome biology. 2015;16:107. https://doi.org/10.1186/s13059-015-0657-6.
Chung YR, Kim HJ, Kim YA, Chang MS, Hwang KT, Park SY. Diversity index as a novel prognostic factor in breast cancer. Oncotarget. 2017;8(57):97114-26. https://doi.org/10.18632/oncotarget.21371.
Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. The Journal of clinical investigation. 2010;120(2):636–44. https://doi.org/10.1172/jci40724.
Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. https://doi.org/10.1016/j.ccr.2007.12.003.
Hosseini H, Obradovic MM, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016; https://doi.org/10.1038/nature20785.
Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nature communications. 2018;9(1):21. https://doi.org/10.1038/s41467-017-02481-5.
Narod SA, Sopik V. Is invasion a necessary step for metastases in breast cancer? Breast Cancer Res Treat. 2018;169(1):9–23. https://doi.org/10.1007/s10549-017-4644-3.
Acknowledgements
The authors would like to acknowledge Drs. Fariba Behbod and Jason I. Herschkowitz for editorial support, as well as Drs. Amanda L. Rinkenbaugh and Abena B. Redwood for critical evaluation and proof reading of this article. This work was supported by the Stand Up To Cancer Laura Ziskin Prize (to HPW) and by the Department of Defense through the Breast Cancer Research Program under Award No. W81XWH-17-1-0077 (to VCS).
Funding
This work was supported by the Stand Up To Cancer Laura Ziskin Prize (to HPW) and by the Department of Defense through the Breast Cancer Research Program under Award No. W81XWH-17-1-0077 (to VCS). Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Sinha, V.C., Piwnica-Worms, H. Intratumoral Heterogeneity in Ductal Carcinoma In Situ: Chaos and Consequence. J Mammary Gland Biol Neoplasia 23, 191–205 (2018). https://doi.org/10.1007/s10911-018-9410-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-018-9410-6