Abstract
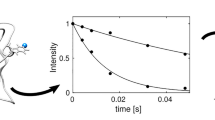
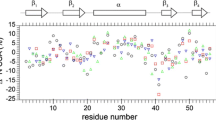
In recent years paramagnetic NMR derived structural constraints have become increasingly popular for the study of biomolecules. Some of these are based on the distance and angular dependences of pseudo contact shifts (PCSs). When modulated by internal motions PCSs also become sensitive reporters on molecular dynamics. We present here an investigation of the domain–domain motion in a two domain protein (PA0128) through time-modulation of PCSs. PA0128 is a protein of unknown function from Pseudomonas aeruginosa (PA) and contains a Zn2+ binding site in the N-terminal domain. When substituted with Co2+ in the binding site, several resonances from the C-terminal domain showed severe line broadening along the 15N dimension. Relaxation compensated CPMG experiments revealed that the dramatic increase in the 15N linewidth came from contributions of chemical exchange. Since several sites with perturbed relaxation are localized to a single β-strand region, and since extracted timescales of motion for the perturbed sites are identical, and since the magnitude of the chemical exchange contributions is consistent with PCSs, the observed rate enhancements are interpreted as the result of concerted domain motion on the timescale of a few milliseconds. Given the predictability of PCS differences and the easy interpretation of the experimental results, we suggest that these effects might be useful in the study of molecular processes occurring on the millisecond to microsecond timescale.






Similar content being viewed by others
Reference
Bax A, Grishaev A (2005) Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol 15:563–570
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2006) 13C-detetced protonless NMR spectroscopy of proteins in solution. Prog Nucl Magn Reson Spectrosc 48:25–45
Bertini I, Bianco CD, Gelis I, Katsaros N, Luchinat C, Parigi G, Peana M, Provenzani A, Zoroddu MA (2004) Experimentally exploring the conformational space sampled by domain reorientation in calmodulin. Proc Natl Acad Sci USA 101:6841–6846
Bertini I, Donaire A, Jimenez B, Luchinat C, Parigi G, Piccioli M, Poggi L (2001a) Paramagnetism-based versus classical constraints: An analysis of the solution structure of Ca Ln calbindin D9k. J Biomol NMR 21:85–98
Bertini I, Janik MBL, Lee YM, Luchinat C, Rosato A (2001b) Magnetic susceptibility tensor anisotropies for a lanthanide ion series in a fixed protein matrix. J Am Chem Soc 123:4181–4188
Bertini I, Longinetti M, Luchinat C, Parigi G, Sgheri L (2002a) Efficiency of paramagnetism-based constraints to determine the spatial arrangement of α-helical secondary structure elements. J Biomol NMR 22:123–136
Bertini I, Luchinat C, Parigi G (2001c) Solution NMR of paramagnetic molecules. Elsevier, Amsterdam
Bertini I, Luchinat C, Parigi G (2002b) Paramagnetic constraints: an aid for quick structure determination of paramagnetic metalloproteins. Concepts Magn Reson 14:259–286
Bertini I, Luchinat C, Piccioli M, Oliver MV, Viezzoli MS (1991) 1H NMR investigation of reduced copper-cobalt superoxide dismutase. Eur Biophys J 20:269–279
Bouvignies G, Bernado P, Meier S, Cho K, Grzesiek S, Bruschweiler R, Blackkedge M (2005) Identification of slow correlated motions in proteins using residual dipolar coupling and hydrogen-bond scalar couplings. Proc Natl Acad Sci USA 102:13885–13890
Clore GM, Omichinski JG, Gronenborn AM (1991) Slow conformational dynamics at the metal coordination site of a zinc finger. J Am Chem Soc 113:4350–4351
Dittmer J, Bodenhausen G (2004) Evidence for slow motion in proteins by multiple refocusing of heteromuclear nitrogen/proton multiple quantum coherences in NMR. J Am Chem Soc 126:1314–1315
Dvoretsky A, Gaponenko V, Rosevear PR (2002) Derivation of structural restraints using thiol-reactive chelator. FEBS Lett 528:189–192
Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D (2005) Intrinsic dynamics of an enzyme underlies catalysis. Nature 438:117–121
Fischer MWF, Losonczi JA, Weaver JL, Prestegard JH (1999) Domain orientation and dynamics in multidomain proteins from residual dipolar couplings. Biochemistry 38:9013–9022
Fruh D, Tolman JR, Bodenhausen G, Zwahlen C (2001) Cross-correlated chemical shift modulation: a signature of slow internal motions in proteins. J Am Chem Soc 123:4810–4816
Gaponenko V, Altieri AS, Li J, Byrd RA (2002) Breaking symmetry in the structure determination of (large) symmetric protein dimers. J Biomol NMR 24:143–148
Hansen DF, Hass MAS, Christensen HM, Ulstrup J, Led JJ (2003) Detection of short-lived transient protein-protein interactions by intermolecular nuclear paramagnetic relaxation: Plastocyanin from Anabaena variabilis. J Am Chem Soc 125:6858–6859
Harper LV, Amann BT, Vinson VK, Berg JM (1993) NMR studies of a cobalt-substituted zinc finger peptide. J Am Chem Soc 115:2577–2580
Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C (2004) Novel techniques for weak alignment of proteins in solution using chemical tags coordinating lanthanide ions. J Biomol NMR 29:339–349
Ishima R, Torchia DA (2000) Protein dynamics from NMR. Nat Struct Biol 7:740–743
Iwahara J, Clore GM (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440:1227–1230
Jensen MR, Lauritzen C, Dahl SW, Pedersen J, Led JJ (2004) Binding ability of a HHP-taged protein toward Ni2+ studied by paramagnetic NMR relaxaton: The possibility of obtaining long-range structure information. J Biomol NMR 29:175–185
Kloiber K, Konrat R (2000) Differential multiple-quantum relaxation arising from cross-correlated time-modulation of isotropic chemical shifts. J Biomol NMR 18:13–42
Korzhnev DM, Neudecker P, Mittermaier A, Orekhov VY, Kay LE (2005) Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: An application to the foldng of a Fyn SH3 domain mutant. J Am Chem Soc 127:15602–15611
Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE (2004) Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature 430:586–590
Kovrigin EL, Loria JP (2006) Characterization of the transition state of functional enzyme dynamics. J Am Chem Soc 128:7724–7725
Loria JP, Rance M, Palmer AG (1999) A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc 121:2331–2332
Lundstrom P, Mulder FAA, Akke M (2005) Correlated dynamics of consecutive residues reveal transient and cooperative unfolding of secondary structure in proteins. Proc Natl Acad Sci USA 102:16984–16989
Ma LX, Jorgensen A-MM, Sorensen GO, Ulstrup J, Led JJ (2000) Elucidation of the paramagnetic R1 relaxation of heteronuclei and protons in Cu(II) plastocyanin from Anabaena variabilis. J Am Chem Soc 122:9473–9485
Ma LX, Led JJ (2000) Determination by high field NMR spectroscopy of the longitudinal electron relaxation rate in Cu(II) plastocyanin form Anabaena variabilis. J Am Chem Soc 122:7823–7824
Majumdar A, Ghose R (2004) Probing slow backbone dynamics in proteins using TROSY-based experiments to detect cross-correlated time-modulation of isotropic chemical shifts. J Biomol NMR 28:213–227
Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG (2000) The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J Am Chem Soc 122:2867–2877
Mispelter J, Momenteau M, Lhoste J-M (1993) Heteronuclear magnetic resonance applications to biological and related paramagnetic molecules. In: Berliner LJ, Reuben J (eds) Biological magnetic resonance: NMR of paramagnetic molecules, vol 12. Plenum Press, New York, pp 299–355
Palmer AG (2004) NMR characterization of the dynamics of biomacromolecules. Chem Rev 104:3623–3640
Pintacuda G, Keniry MA, Huber T, Park AY, Dixon NE, Otting G (2004a) Fast structure-based assignment of 15N HSQC spectra of selectively 15N-labeled paramagnetic proteins. J Am Chem Soc 126:2963–2970
Pintacuda G, Moshref A, Leonchiks A, Sharipo A, Otting G (2004b) Site-specific labeling with a metal chelator for protein-structure refinement. J Biomol NMR 29:351–361
Prestegard JH, Bougault CM, Kishore AI (2004) Residual dipolar couplings in structure determination of biomolecules. Chem Rev 104:3519–3540
Rodriguez-Castaneda F, Haberz P, Leonov A, Griesinger C (2006) Pamagnetic tagging of diamagnetic proteins for solution NMR. Magn Reson Chem 44:S10–S16
Tang C, Iwahara J, Clore GM (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444:383–386
Tolman JR, Flanagan JM, Kennesy MA, Prestegard JH (1995) Nuclear magnetic dipole interactions in field-oriented proteins – information for structure determination in solution. Proc Natl Acad Sci USA 92:9279–9283
Tolman JR, Flanagan JM, Kennesy MA, Prestegard JH (1997) NMR evidence for slow collective motions in cyanometmyoglobin. Nat Struct Biol 4:292–297
Volkov AN, Worrall JAR, Holtzmann E, Ubbink M (2006) Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci 103:18945–18950
Wohnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H (2003) Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J Am Chem Soc 125:13338–13339
Acknowledgement
This work is supported by NIH grants GM074958, GM33225 and the Ontario Research and Development Fund. We thank professor Mark Rance at University of Cincinnati for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, X., Srisailam, S., Yee, A.A. et al. Domain–domain motions in proteins from time-modulated pseudocontact shifts. J Biomol NMR 39, 53–61 (2007). https://doi.org/10.1007/s10858-007-9174-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-007-9174-6