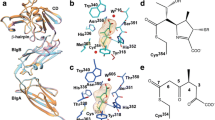
Abstract
Tuberculosis remains a dreadful disease that has claimed many human lives worldwide and elimination of the causative agent Mycobacterium tuberculosis also remains elusive. Multidrug-resistant TB is rapidly increasing worldwide; therefore, there is an urgent need for improving the current antibiotics and novel drug targets to successfully curb the TB burden. l,d-Transpeptidase 2 is an essential protein in Mtb that is responsible for virulence and growth during the chronic stage of the disease. Both d,d- and l,d-transpeptidases are inhibited concurrently to eradicate the bacterium. It was recently discovered that classic penicillins only inhibit d,d-transpeptidases, while l,d-transpeptidases are blocked by carbapenems. This has contributed to drug resistance and persistence of tuberculosis. Herein, a hybrid two-layered ONIOM (B3LYP/6-31G+(d): AMBER) model was used to extensively investigate the binding interactions of LdtMt2 complexed with four carbapenems (biapenem, imipenem, meropenem, and tebipenem) to ascertain molecular insight of the drug-enzyme complexation event. In the studied complexes, the carbapenems together with catalytic triad active site residues of LdtMt2 (His187, Ser188 and Cys205) were treated at with QM [B3LYP/6-31+G(d)], while the remaining part of the complexes were treated at MM level (AMBER force field). The resulting Gibbs free energy (ΔG), enthalpy (ΔH) and entropy (ΔS) for all complexes showed that the carbapenems exhibit reasonable binding interactions towards LdtMt2. Increasing the number of amino acid residues that form hydrogen bond interactions in the QM layer showed significant impact in binding interaction energy differences and the stabilities of the carbapenems inside the active pocket of LdtMt2. The theoretical binding free energies obtained in this study reflect the same trend of the experimental observations. The electrostatic, hydrogen bonding and Van der Waals interactions between the carbapenems and LdtMt2 were also assessed. To further examine the nature of intermolecular interactions for carbapenem–LdtMt2 complexes, AIM and NBO analysis were performed for the QM region (carbapenems and the active residues of LdtMt2) of the complexes. These analyses revealed that the hydrogen bond interactions and charge transfer from the bonding to anti-bonding orbitals between catalytic residues of the enzyme and selected ligands enhances the binding and stability of carbapenem–LdtMt2 complexes.
Graphical abstract
The two-layered ONIOM (B3LYP/6-31+G(d): Amber) model was used to evaluate the efficacy of FDA approved carbapenems antibiotics towards LdtMt2.








Similar content being viewed by others
References
World Health Organization (WHO) (2017) Global tuberculosis report, 2016 [cited 16 January 2018]. http://www.who.int/tb/publications/global_report/en/
Cegielski JP (2010) Extensively drug-resistant tuberculosis:“there must be some kind of way out of here”. Clin Infect Dis 50:S195–S200
Cole ST, Riccardi G (2011) New tuberculosis drugs on the horizon. Curr Opin Microbiol 14:570–576
Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G (2006) Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575–1580
Organization WH (2014) World Health Organization global tuberculosis report. World Health Organization, Geneva
Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G (2010) The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 16:466–469
Mainardi J-L, Villet R, Bugg TD, Mayer C, Arthur M (2008) Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev 32:386–408
Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi J-L (2008) The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J Bacteriol 190:4360–4366
Erdemli SB, Gupta R, Bishai WR, Lamichhane G, Amzel LM, Bianchet MA (2012) Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of l,d-transpeptidase 2. Structure 20:2103–2115
Li WJ, Li DF, Hu YL, Zhang XE, Bi LJ, Wang DC (2013) Crystal structure of l,d-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis. Cell Res 23:728–731
Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA (2011) Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960
Wietzerbin J, Das BC, Petit JF, Lederer E, Leyh-Bouille M, Ghuysen JM (1974) Occurrence of d-alanyl-(d)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry 13:3471–3476
Böth D, Steiner EM, Stadler D, Lindqvist Y, Schnell R, Schneider G (2013) Structure of LdtMt2, an l,d-transpeptidase from Mycobacterium tuberculosis. Acta Crystallogr D 69:432–441
Cordillot M, Dubée V, Triboulet S, Dubost L, Marie A, Hugonnet J-E, Arthur M, Mainardi J-L (2013) In vitro cross-linking of Mycobacterium tuberculosis peptidoglycan by l,d-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrob Agents Chemother 57:5940–5945
Correale S, Ruggiero A, Capparelli R, Pedone E, Berisio R (2013) Structures of free and inhibited forms of the l,d-transpeptidase LdtMt1 from Mycobacterium tuberculosis. Acta Crystallogr D 69:1697–1706
Dubée V, Triboulet S, Mainardi J-L, Ethève-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet J-E, Arthur M (2012) Inactivation of Mycobacterium tuberculosis l,d-transpeptidase LdtMt1 by carbapenems and cephalosporins. Antimicrob Agents Chemother 56:4189–4195
Schoonmaker MK, Bishai WR, Lamichhane G (2014) Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to β-lactams. J Bacteriol 196:1394–1402
Kim HS, Im KJ, Yoon HN, An JY, Yoon DR, Kim HJ, Min JY, Kim HK, Lee SJ, Han JY, Suh BW (2013) Structural basis for the inhibition of Mycobacterium tuberculosis l,d-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains. Acta Crystallogr D D69:420–431
Nizami B, Sydow D, Wolber G, Honarparvar B (2016) Molecular insight on the binding of NNRTI to K103N mutated HIV-1 RT: molecular dynamics simulations and dynamic pharmacophore analysis. Mol BioSyst 12:3385–3395
Honarparvar B, Pawar SA, Alves CN, Lameira J, Maguire GE, Silva JRA, Govender T, Kruger HG (2015) Pentacycloundecane lactam vs lactone norstatine type protease HIV inhibitors: binding energy calculations and DFT study. J Biomed Sci 22:15
Honarparvar B, Govender T, Maguire GE, Soliman ME, Kruger HG (2013) Integrated approach to structure-based enzymatic drug design: molecular modeling, spectroscopy, and experimental bioactivity. Chem Rev 114:493–537
Silva JRA, Bishai WR, Govender T, Lamichhane G, Maguire GE, Kruger HG, Lameira J, Alves CN (2016) Targeting the cell wall of Mycobacterium tuberculosis: a molecular modeling investigation of the interaction of imipenem and meropenem with l,d-transpeptidase 2. J Biomol Struct Dyn 34:304–317
Fakhar Z, Govender T, Maguire GEM, Lamichhane G, Walker RC, Kruger HG, Honarparvar B (2017) Differential flap dynamics in l,d-transpeptidase 2 from Mycobacterium tuberculosis revealed by molecular dynamics. Mol BioSyst 13:1223–1234
Svensson M, Humbel S, Froese RD, Matsubara T, Sieber S, Morokuma K (1996) ONIOM: a multilayered integrated MO + MM method for geometry optimizations and single point energy predictions. A test for Diels-Alder reactions and Pt(P(t–Bu)3)2 + H2 oxidative addition. J Phys Chem A 100:19357–19363
Dapprich S, Komáromi I, Byun KS, Morokuma K, Frisch MJ (1999) A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. Comput Theor Chem 461:1–21
Vreven T, Morokuma K (2000) On the application of the IMOMO (integrated molecular orbital + molecular orbital) method. J Comput Chem 21:1419–1432
Karadakov PB, Morokuma K (2000) ONIOM as an efficient tool for calculating NMR chemical shielding constants in large molecules. Chem Phys Lett 317:589–596
Banáš P, Jurečka P, Walter NG, Šponer J, Otyepka M (2009) Theoretical studies of RNsA catalysis: hybrid QM/MM methods and their comparison with MD and QM. Methods 49:202–216
Morokuma K (2009) Theoretical studies of structure, function and reactivity of molecules—a personal account. Proc Jpn Acad Ser B 85:167–182
Masessras F, Morokuma K (1995) IMOMM: a new integrated ab initio + molecular mechanics geometry optimization scheme of equilibrium structures and transition states. J Comp Chem 16:1170–1179
Zheng F, Zhan C-G (2008) Rational design of an enzyme mutant for anti-cocaine therapeutics. J Comput-Aided Mol Des 22:661–671
Ruangpornvisuti V (2004) Recognition of carboxylate and dicarboxylates by azophenol–thiourea derivatives: a theoretical host–guest investigation. Comput Theor Chem 686:47–55
Samanta PN, Das KK (2016) Prediction of binding modes and affinities of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide inhibitors to the carbonic anhydrase receptor by docking and ONIOM calculations. J Mol Graph Model 63:38–48
Promsri S, Chuichay P, Sanghiran V, Parasuk V, Hannongbua S (2005) Molecular and electronic properties of HIV-1 protease inhibitor C 60 derivatives as studied by the ONIOM method. Comput Theor Chem 715:47–53
Bader R, Molecules AI (1990) A quantum theory. Clarendon, Oxford
Reed AE, Curtiss LA, Weinhold F (1988) Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem Rev 88:899–926
Li W-J, Li D-F, Hu Y-L, Zhang X-E, Bi L-J, Wang D-C (2013) Crystal structure of l,d-transpeptidase LdtMt2 in complex with meropenem reveals the mechanism of carbapenem against Mycobacterium tuberculosis. Cell Res 23:728
The PyMOL Molecular Graphics System, V. S., LLC
Case D, Darden T, Cheatham III TE, Simmerling T, Wang C, Duke J, Luo R, Crowley R, Walker M, R., and Zhang W (2008) AMBER, version 10. University of California, San Francisco
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Discipl Protein Biochem 65:712–725
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Harvey MJ, de Fabritiis G (2009) An implementation of the smooth particle Mesh Ewald method on GPU hardware. J Chem Theory Comput 5:2371–2377
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of N-alkanes. J Comput Phys 23:327–341
Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins Discipl Protein Biochem 61:704–721
Frisch M, Trucks G, Schlegel H, Scuseria G, Robb M, Cheeseman J, Scalmani G, Barone V, Mennucci B, Petersson G (2009) Gaussian 2009 (see supporting information for full citation)
Kapp J, Remko M, Schleyer PvR (1996) H2XO and (CH3)2XO Compounds (X = C, Si, Ge, Sn, Pb): double bonds vs carbene-like structures can the metal compounds exist at all? J Am Chem Soc 118:5745–5751
Remko M, Walsh OA, Richards WG (2001) Theoretical study of molecular structure, tautomerism, and geometrical isomerism of moxonidine: Two-layered ONIOM calculations. J Phys Chem A 105:6926–6931
Johnson BG, Gill PM, Pople JA (1993) The performance of a family of density functional methods. J Chem Phys 98:5612–5626
Humbel S, Sieber S, Morokuma K (1996) The IMOMO method: integration of different levels of molecular orbital approximations for geometry optimization of large systems: test for n-butane conformation and SN2 reaction: RCl + Cl–. J Chem Phys 105:1959–1967
Kohn W, Becke AD, Parr RG (1996) Density functional theory of electronic structure. J Phys Chem A 100:12974–12980
Neumann R, Nobes RH, Handy NC (1996) Exchange functionals and potentials. Mol Phys 87:1–36
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37:785
Hariharan PC, Pople JA (1973) The influence of polarization functions on molecular orbital hydrogenation energies. Theoretica chimica acta 28:213–222
Rassolov VA, Pople JA, Ratner MA, Windus TL (1998) 6-31G* basis set for atoms K through Zn. J Chem Phys 109:1223–1229
Vreven T, Byun KS, Komáromi I, Dapprich S, Montgomery JA, Morokuma K, Frisch MJ (2006) Combining quantum mechanics methods with molecular mechanics methods in ONIOM. J Chem Theory Comput 2:815–826
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786
Panigrahi SK (2008) Strong and weak hydrogen bonds in protein-ligand complexes of kinases: a comparative study. Amino Acids 34:617–633
Desiraju GR (1996) The C–H⋯O hydrogen bond: structural implications and supramolecular design. Acc Chem Res 29:441–449
Panigrahi SK, Desiraju GR (2007) Strong and weak hydrogen bonds in the protein–ligand interface. Proteins Discipl Protein Biochem 67:128–141
Dassault Systèmes BIOVIA, Release DSME (2017) San Diego: Dassault Systèmes, 2016
Popelier P, Bader R (1992) The existence of an intramolecular C–H–O hydrogen bond in creatine and carbamoyl sarcosine. Chem Phys Lett 189:542–548
Bianchet MA, Pan YH, Basta LAB, Saavedra H, Lloyd EP, Kumar P, Mattoo R, Townsend CA, Lamichhane G (2017) Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem. BMC Biochem 18:8
Lu Y, Wang Y, Xu Z, Yan X, Luo X, Jiang H, Zhu W (2009) C–X⋯H contacts in biomolecular systems: how they contribute to protein–ligand binding affinity. J Phys Chem B 113:12615–12621
Patil R, Das S, Stanley A, Yadav L, Sudhakar A, Varma AK (2010) Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS ONE 5:e12029
Bissantz C, Kuhn B, Stahl M (2010) A medicinal chemist’s guide to molecular interactions. J Med Chem 53:5061–5084
Parthasarathi R, Amutha R, Subramanian V, Nair BU, Ramasami T (2004) Bader’s and reactivity descriptors’ analysis of DNA base pairs. J Phys Chem A 108:3817–3828
Biegler-Konig F, Schonbohm J, Bayles D (2001) Software news and updates-AIM2000-A program to analyze and visualize atoms in molecules. Wiley, New York
Popelier P (1998) Characterization of a dihydrogen bond on the basis of the electron density. J Phys Chem A 102:1873–1878
Bader RF (1990) Atoms in molecules: a quantum theory. International series of monographs on chemistry, 22, Oxford University Press, Oxford Henkelman G, Arnaldsson A, Jónsson H (2006) A fast and robust algorithm for Bader decomposition of charge density. Comput Mater Sci 36:354–360
Nazari F, Doroodi Z (2010) The substitution effect on heavy versions of cyclobutadiene. Int J Quantum Chem 110:1514–1528
Mosapour Kotena Z, Behjatmanesh–Ardakani R, Hashim R (2014) AIM and NBO analyses on hydrogen bonds formation in sugar-based surfactants (α/β-d-mannose and n-octyl-α/β-d-mannopyranoside): a density functional theory study. Liq Cryst 41:784–792
Acknowledgements
We thank the College of Health Sciences (CHS), Aspen Pharmacare, MRC and NRF for financial support. We are also grateful to the CHPC (http://www.chpc.ac.za) and UKZN cluster for computational resources.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ntombela, T., Fakhar, Z., Ibeji, C.U. et al. Molecular insight on the non-covalent interactions between carbapenems and l,d-transpeptidase 2 from Mycobacterium tuberculosis: ONIOM study. J Comput Aided Mol Des 32, 687–701 (2018). https://doi.org/10.1007/s10822-018-0121-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0121-2