Abstract
Mating-type (MAT) locus-specific single nucleotide polymorphisms (SNPs) have been shown to be sufficient for conventional PCR-based differentiation of Pyrenophora teres f. teres (Ptt) and P. teres f. maculata (Ptm), the cause of the net and spot form, respectively, of barley net blotch. Here, we report the cloning and characterization of the MAT locus from 10 California isolates that cause atypical blotch symptoms on barley. Analysis of the full-length nucleotide sequences of one MAT1-1 (1,993 bp) and nine MAT1-2 (2,149 or 2,161 bp) idiomorphs revealed high (98–99 %) similarity to those of Ptt isolates. However, distinct SNP patterns were identified in the newly cloned MAT idiomorphs. Two new MAT1-2-specific SNPs were found to be conserved in one Australia and eight California isolates that all cause similar atypical blotch symptoms. Phylogenetic analysis indicated that all 10 California isolates form a separate branch (or clade) within the Ptt group, except for one that appears to be ancestral to both Ptt and Ptm. PCR primers designed based on the identified SNP patterns were used successfully to differentiate each atypical isolate from highly virulent forms. This study extends our previous work and, taken together, the results demonstrate that the genetic variation at the MAT locus correlates with variation in the form-specific disease phenotype, and that MAT-specific SNPs can serve as reliable and convenient markers for subspecies level differentiation in P. teres, an economically important plant-pathogenic ascomycete (http://en.wikipedia.org/wiki/Single_nucleotide_polymorphism).






Similar content being viewed by others
References
Abu Qamar, M., Liu, Z. H., Faris, J. D., Chao, S., Edwards, M. C., Lai, Z., et al. (2008). A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theoretical and Applied Genetics, 117, 1261–1270.
Aitken, N., Smith, S., Schwarz, C., & Morin, P. A. (2004). Single nucleotide polymorphism (SNP) discovery in mammals: a targeted-gene approach. Molecular Ecology, 13, 1423–1431.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Andrie, R. M., Schoch, C. L., Hedges, R., Spatafora, J. W., & Ciuffetti, L. M. (2008). Homologs of ToxB, a host-selective toxin gene from Pyrenophora tritici-repentis, are present in the genome of sister-species Pyrenophora bromi and other members of the Ascomycota. Fungal Genetics and Biology, 45, 363–377.
Campbell, G. F., Crows, P. W., & Lucas, J. A. (1999). Pyrenophora teres f. maculata, the cause of Pyrenophora leaf spot of barley in South Africa. Mycological Research, 103, 257–267.
Chagne, D., Batley, J., Edwards, D., & Forster, J. W. (2007). Single nucleotide polymorphism genotyping in plants. In N. C. Oraguzie, E. H. A. Rikkerink, S. E. Gardinerand, & H. N. De Silva (Eds.), Association mapping in plants (pp. 77–94). New York: Springer LLC.
Chilvers, M. I., du Toit, L. J., Akamatsu, H., & Peever, T. L. (2007). A real-time, quantitative PCR seed assay for Botrytis spp. that cause neck rot of onion. Plant Disease, 91, 599–608.
Debuchy, R., & Turgeon, B. G. (2006). Mating-type structure, evolution, and function in Euascomycetes. In U. Kues & R. Fischer (Eds.), The mycota, vol I (pp. 293–323). Berlin: Springer.
Ellwood, S. R., Liu, Z., Syme, R. A., Lai, Z., Hane, J. K., Keiper, F., et al. (2010). A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres. Genome Biology, 11, R109.
Felsenstein, J. (1989). Mathematics vs. evolution: mathematical evolutionary theory. Science, 246, 941–942.
Fournier, A., Widmer, F., & Enkerli, J. (2010). Development of a single-nucleotide polymorphism (SNP) assay for genotyping of Pandora neoaphidis. Fungal Biology, 114, 498–506.
Friesen, T. L., Faris, J. D., Lai, Z., & Steffenson, B. J. (2006). Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled-haploid barley population. Genome, 49, 855–859.
Grewal, T. S., Rossnagel, B. G., Pozniak, C. J., & Scoles, G. J. (2008). Mapping quantitative trait loci associated with barley net blotch resistance. Theoretical and Applied Genetics, 116, 529–539.
Jonsson, R., Sall, T., & Bryngelsson, T. (2000). Genetic diversity for random amplified polymorphic DNA (RAPD) markers in two Swedish populations of Pyrenophora teres. Canadian Journal of Plant Pathology, 22, 258–264.
Kristensen, R., Berdal, K. G., & Holst-Jensen, A. (2007). Simultaneous detection and identification of trichothecene- and moniliformin-producing Fusarium species based on multiplex SNP analysis. Journal of Applied Microbiology, 102, 1071–1081.
Lai, Z., Faris, J. D., Weiland, J. J., Steffenson, B. J., & Friesen, T. L. (2007). Genetic mapping of Pyrenophora teres f. teres genes conferring avirulence on barley. Fungal Genetics and Biology, 44, 323–329.
Leisova, L., Kucera, L., Minarikova, V., & Ovesna, J. (2005). AFLP-based PCR markers that differentiate spot and net forms of Pyrenophora teres. Plant Pathology, 54, 66–73.
Lepoint, P., Renard, M. E., Legreve, A., Duveiller, E., & Maraite, H. (2010). Genetic diversity of the mating type and toxin production genes in Pyrenophora tritici-repentis. Phytopathology, 100, 474–483.
Liu, Z. H., & Friesen, T. L. (2010). Identification of Pyrenophora teres f. maculata, causal agent of spot type net blotch of barley in North Dakota. Plant Disease, 94, 480.
Liu, Z. H., Ellwood, S. R., Oliver, R. P., & Friesen, T. L. (2011). Pyrenophora teres: profile of an increasingly damaging barley pathogen. Molecular Plant Pathology, 12, 1–19.
Lu, S., Platz, G. J., Edwards, M. C., & Friesen, T. L. (2010). Mating type (MAT) locus-specific PCR markers for differentiation of Pyrenophora teres f. teres and P. teres f. maculata, the causal agents of barley net blotch. Phytopathology, 100, 1298–1306.
Martin, T., Lu, S., van Tilbeurgh, H., Ripoll, D. R., Dixelius, C., Turgeon, B. G., & Debuchy, R. (2010). Tracing the origin of the fungal sex α1 domain places its ancestor in the HMG-box superfamily: implication for fungal mating-type evolution. PLoS One, 5(12), e15199.
McLean, M. S., Howlett, B. J., & Hollaway, G. J. (2009). Epidemiology and control of spot form of net blotch (Pyrenophora teres f. maculata) of barley: a review. Crop Pasture Science, 60, 499–499.
O’Donnell, K., Ward, T. J., Geiser, D. M., Corby Kistler, H., & Aoki, T. (2004). Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology, 41, 600–623.
Rafalski, A. (2002). Applications of single nucleotide polymorphisms in crop genetics. Current Opinion in Plant Biology, 5, 94–100.
Rau, D., Maier, F. J., Papa, R., Brown, A. H., Balmas, V., Saba, E., et al. (2005). Isolation and characterization of the mating-type locus of the barley pathogen Pyrenophora teres and frequencies of mating-type idiomorphs within and among fungal populations collected from barley landraces. Genome, 48, 855–869.
Rau, D., Attene, G., Brown, A. H., Nanni, L., Maier, F. J., Balmas, V., et al. (2007). Phylogeny and evolution of mating-type genes from Pyrenophora teres, the causal agent of barley “net blotch” disease. Current Genetics, 51, 377–392.
Scott, D. B. (1991). Identity of Pyrenophora isolates causing net-type and spot-type lesions on barley. Mycopathologia, 116, 29–35.
Serenius, M., Manninen, O., Wallwork, H., & Williams, K. (2007). Genetic differentiation in Pyrenophora teres populations measured with AFLP markers. Mycological Research, 111, 213–223.
Smedegård-Petersen, V. (1971). Pyrenophora teres fsp. maculata f. nov and Pyrenophora teres f. teres on barley in Denmark. Yearbook of Royal Veterinary and Agricultural University (Copenhagen) 1971, pp 124–144.
Syvanen, A. C. (2001). Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Reviews Genetics, 2, 930–942.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., & Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882.
Turgeon, B. G. (1998). Application of mating type gene technology to problems in fungal biology. Annual Review of Phytopathology, 36, 115–137.
Turgeon, B. G., & Yoder, O. C. (2000). Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genetics and Biology, 31, 1–5.
Wang, D. G., Fan, J. B., Siao, C. J., Berno, A., Young, P., Sapolsky, R., et al. (1998). Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082.
Yun, S. H., Arie, T., Kaneko, I., Yoder, O. C., & Turgeon, B. G. (2000). Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genetics and Biology, 31, 7–20.
Acknowledgments
We thank Kelsey Dunnell and Danielle Holmes for technical assistance. This study was supported by USDA-ARS CRIS projects 5442-21000-033-00D and 5442-22000-043-00D and the NIFA-AFRI Coordinated Agricultural Project (CAP) Competitive Grant no. 2011-68002-30029.
Author information
Authors and Affiliations
Corresponding author
Additional information
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
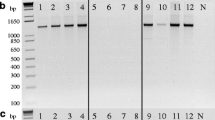
Nucleotide sequence data reported are available in the GenBank databases under the accession numbers JN980080 (for MAT1-1 idiomorph of isolate CAWB05-Pt-5), JN980081 to JN980088 (for MAT1-2 idiomorphs of CAWB1 to 5, CAWB05-Pt-2 to Pt-4, respectively) and JN980089 (for MAT1-2 idiomorph of CAWB05-Pt-1).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
Nucleotide sequence alignments of internal transcribed spacer (ITS) regions of the 10 California isolates identified in this study and representative isolates of P. teres f. teres (Ptt), P. teres f. maculata (Ptm) and P. graminea (Pg) reported in previous studies (Andrie et al. 2008; Liu and Friesen 2010). The ITS regions of the 10 California isolates that include 18S ribosomal RNA gene (partial sequence), internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, and 28S ribosomal RNA gene (partial sequence) were PCR-amplified with primers 5′-TCCGTAGGTGAACCTGCGG-3′ and 5′-TCCTCCGCTTATTGATATGC-3′. Shown here are the sequences corresponding to nucleotide positions 7 to 476 of GenBank accession number GU014819 (Ptt isolate 0-1, Liu and Friesen 2010). Note that only one P. graminea-specific single nucleotide polymorphism (SNP) site was identified (triangle). The variations found in the two short stretches of T-rich sequences (asterisks) are not form- or species-specific and are likely due to PCR or sequencing errors. (DOCX 662 kb)
Supplemental Figure 2
Phylogenetic relationships of the glyceraldehyde-3-phosphate dehydrogenase-like (GPD) genes of P. teres isolates. A 570-bp genomic sequence of the GPD gene was PCR-amplified from the 10 California isolates, the Australia isolate SNB172i, and 16 other P. teres f. teres (Ptt) and nine P. teres f. maculata (Ptm) isolates (Lu et al. 2010). PCR primers (5′-CGTATCGTCTTCCGCAAC -3′ and 5′-TTGGAGAGCACCTCAATGT -3′) were designed to GenBank accession number EF513236 (Ptt isolate ND, Andrie et al. 2008). The amplified sequences were aligned using the CLUSTALX program (Thompson et al. 1997) and the neighbor joining tree was constructed with the PHYLIP 3.61 package (http://evolution.genetics.washington.edu/phylip.html; Felsenstein 1989) and drawn using TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Bootstrapping values (>50 %, from 1,000 replicates) are indicated on the branches. A part of the sequence alignment was given below to show the identified polymorphic sites (triangles). Nucleotide sequences of the PCR-amplified GPD genes have been deposited into the GenBank database under accession numbers JQ837841 to JQ837876. (DOCX 198 kb)
Supplemental Figure 3
Nucleotide sequence alignments of MAT1-2 idiomorphs of the nine California isolates and other P. teres f. teres isolates identified from previous studies. Gray bars on the top indicate the highly conserved sequences flanking the MAT idiomorphs. Triangles indicate the positions of conserved single nucleotide polymorphisms (SNPs) with the corresponding nucleotide and the positions indicated at the top. Black triangles indicate the six SNPs used to differentiate typical isolates of P. teres f. teres from P. teres f. maculata. White triangles indicate the two newly identified SNPs conserved among the eight California isolates and the Australia isolate SNB172i (brackets). Gray triangle indicates a less conserved SNP which is found in six California isolates (CAWB3 to 5 and CAWB05-Pt-2 to Pt-4). Asterisk indicates the positions of a 12-bp deletion in CAWB05-Pt-1. Start and stop codons of the MAT1-2-1 open reading frames are underlined. Intron sequences are boxed. GenBank accession numbers: CAWB1 to 5, JN980081 to JN980085; CAWB05-Pt-2 to Pt-4, JN980086 to JN980088; CAWB05-Pt-1, JN980089; 0-1, AY950585 (Ellwood et al. 2010); BrPteres, HM121999; Jpt0101, HM121998; LDNH04Pt1, HM121998; NB077, HM122000; SNB172i, HM122001 (Lu et al. 2010). (DOCX 1.08 kb)
Rights and permissions
About this article
Cite this article
Lu, S., Edwards, M.C. & Friesen, T.L. Genetic variation of single nucleotide polymorphisms identified at the mating type locus correlates with form-specific disease phenotype in the barley net blotch fungus Pyrenophora teres . Eur J Plant Pathol 135, 49–65 (2013). https://doi.org/10.1007/s10658-012-0064-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-012-0064-8