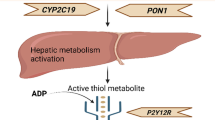
Abstract
Purpose
Describe CYP2C19 sequencing results in the largest series of clopidogrel-treated cases with stent thrombosis (ST), the closest clinical phenotype to clopidogrel resistance. Evaluate the impact of CYP2C19 genetic variation detected by next-generation sequencing (NGS) with comprehensive annotation and functional studies.
Methods
Seventy ST cases on clopidogrel identified from the PLATO trial (n = 58) and Mayo Clinic biorepository (n = 12) were matched 1:1 with controls for age, race, sex, diabetes mellitus, presentation, and stent type. NGS was performed to cover the entire CYP2C19 gene. Assessment of exonic variants involved measuring in vitro protein expression levels. Intronic variants were evaluated for potential splicing motif variations.
Results
Poor metabolizers (n = 4) and rare CYP2C19*8, CYP2C19*15, and CYP2C19*11 alleles were identified only in ST cases. CYP2C19*17 heterozygote carriers were observed more frequently in cases (n = 29) than controls (n = 18). Functional studies of CYP2C19 exonic variants (n = 11) revealed 3 cases and only 1 control carrying a deleterious variant as determined by in vitro protein expression studies. Greater intronic variation unique to ST cases (n = 169) compared with controls (n = 84) was observed with predictions revealing 13 allele candidates that may lead to a potential disruption of splicing and a loss-of-function effect of CYP2C19 in ST cases.
Conclusion
NGS detected CYP2C19 poor metabolizers and paradoxically greater number of so-called rapid metabolizers in ST cases. Rare deleterious exonic variation occurs in 4%, and potentially disruptive intronic alleles occur in 16% of ST cases. Additional studies are required to evaluate the role of these variants in platelet aggregation and clopidogrel metabolism.



Similar content being viewed by others
References
Doyle B, Rihal CS, O’Sullivan CJ, Lennon RJ, Wiste HJ, Bell M, et al. Outcomes of stent thrombosis and restenosis during extended follow-up of patients treated with bare-metal coronary stents. Circulation. 2007;116(21):2391–8.
Cayla G, Hulot JS, O'Connor SA, Pathak A, Scott SA, Gruel Y, et al. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. Jama. 2011;306(16):1765–74.
Pereira NL, Rihal CS, So DYF, Rosenberg Y, Lennon RJ, Mathew V, et al. Clopidogrel pharmacogenetics. Circ Cardiovasc Interv. 2019;12(4):e007811.
Bernard G, Chouery E, Putorti ML, Tetreault M, Takanohashi A, Carosso G, et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(3):415–23.
Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–62.
Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–75.
Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304(16):1821–30.
Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849–57.
Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376(9749):1320–8.
Olson JE, Ryu E, Johnson KJ, Koenig BA, Maschke KJ, Morrisette JA, et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88(9):952–62.
Andell P, James SK, Cannon CP, Cyr DD, Himmelmann A, Husted S, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes and chronic obstructive pulmonary disease: an analysis from the Platelet Inhibition and Patient Outcomes (PLATO) Trial. J Am Heart Assoc. 2015;4(10):e002490.
St Sauver JL, Grossardt BR, Yawn BP, Melton LJ 3rd, Pankratz JJ, Brue SM, et al. Data resource profile: the Rochester Epidemiology Project (REP) medical records-linkage system. Int J Epidemiol. 2012;41(6):1614–24.
Gerber Y, Weston SA, Enriquez-Sarano M, Jaffe AS, Manemann SM, Jiang R, et al. Contemporary risk stratification after myocardial infarction in the community: performance of scores and incremental value of soluble suppression of tumorigenicity-2. J Am Heart Assoc. 2017;6(10):e005958.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 0 1–0 33.
Matreyek KA, Stephany JJ, Fowler DM. A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Res. 2017;45(11):e102.
Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 2018;50(6):874–82.
Wrenbeck EE, Klesmith JR, Stapleton JA, Adeniran A, Tyo KE, Whitehead TA. Plasmid-based one-pot saturation mutagenesis. Nat Methods. 2016;13(11):928–30.
Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15(17):7155–74.
Zhang MQ. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7(5):919–32.
Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2–3):377–94.
Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29(5):1185–90.
Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4(3):311–23.
Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, et al. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015;25(2):290–303.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67.
Lin H, Hargreaves KA, Li R, Reiter JL, Wang Y, Mort M, et al. RegSNPs-intron: a computational framework for predicting pathogenic impact of intronic single nucleotide variants. Genome Biol. 2019;20(1):254.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7.
Takahashi M, Saito T, Ito M, Tsukada C, Katono Y, Hosono H, et al. Functional characterization of 21 CYP2C19 allelic variants for clopidogrel 2-oxidation. Pharmacogenomics J. 2015;15(1):26–32.
Blaisdell J, Mohrenweiser H, Jackson J, Ferguson S, Coulter S, Chanas B, et al. Identification and functional characterization of new potentially defective alleles of human CYP2C19. Pharmacogenetics. 2002;12(9):703–11.
Holmes DR Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol. 2010;56(4):321–41.
Pereira NL, Geske JB, Mayr M, Shah SH, Rihal CS. Pharmacogenetics of clopidogrel: an unresolved issue. Circ Cardiovasc Genet. 2016;9(2):185–8.
Pereira NL, Sargent DJ, Farkouh ME, Rihal CS. Genotype-based clinical trials in cardiovascular disease. Nat Rev Cardiol. 2015;12(8):475–87.
Pereira NL, Stewart AK. Clinical implementation of cardiovascular pharmacogenomics. Mayo Clin Proc. 2015;90(6):701–4.
Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014;312(18):1870–9.
Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–23.
Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121(4):512–8.
Gladding P, Webster M, Zeng I, Farrell H, Stewart J, Ruygrok P, et al. The pharmacogenetics and pharmacodynamics of clopidogrel response: an analysis from the PRINC (Plavix response in coronary intervention) trial. JACC Cardiovasc Interv. 2008;1(6):620–7.
Gurbel PA, Shuldiner AR, Bliden KP, Ryan K, Pakyz RE, Tantry US. The relation between CYP2C19 genotype and phenotype in stented patients on maintenance dual antiplatelet therapy. Am Heart J. 2011;161(3):598–604.
Sorich MJ, Polasek TM, Wiese MD. Systematic review and meta-analysis of the association between cytochrome P450 2C19 genotype and bleeding. Thromb Haemost. 2012;108(1):199–200.
Pedersen RS, Nielsen F, Stage TB, Vinholt PJ, el Achwah AB, Damkier P, et al. CYP2C19*17 increases clopidogrel-mediated platelet inhibition but does not alter the pharmacokinetics of the active metabolite of clopidogrel. Clin Exp Pharmacol Physiol. 2014;41(11):870–8.
Wang H, An N, Wang H, Gao Y, Liu D, Bian T, et al. Evaluation of the effects of 20 nonsynonymous single nucleotide polymorphisms of CYP2C19 on S-mephenytoin 4′-hydroxylation and omeprazole 5′-hydroxylation. Drug Metab Dispos. 2011;39(5):830–7.
Devarajan S, Moon I, Ho MF, Larson NB, Neavin DR, Moyer AM, et al. Pharmacogenomic next-generation DNA sequencing: lessons from the identification and functional characterization of variants of unknown significance in CYP2C9 and CYP2C19. Drug Metab Dispos. 2019;47(4):425–35.
Quax TE, Claassens NJ, Soll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59(2):149–61.
Stein KC, Frydman J. The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J Biol Chem. 2019;294(6):2076–84.
Jo BS, Choi SS. Introns: the functional benefits of introns in genomes. Genomics Inform. 2015;13(4):112–8.
Chaudhry AS, Prasad B, Shirasaka Y, Fohner A, Finkelstein D, Fan Y, et al. The CYP2C19 intron 2 branch point SNP is the ancestral polymorphism contributing to the poor metabolizer phenotype in livers with CYP2C19*35 and CYP2C19*2 alleles. Drug Metab Dispos. 2015;43(8):1226–35.
Brown SA, Pereira N. Pharmacogenomic impact of CYP2C19 variation on clopidogrel therapy in precision cardiovascular medicine. J Pers Med. 2018;8(1):E8.
Funding
This study was supported by NIH/NHLBI grant U01 HL128606 to N.L.P. PLATO study was funded by AstraZeneca.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The protocol was reviewed by the IRB and approval was obtained for this study.
Disclaimer
The sponsor had no role in data analysis, interpretation, or manuscript writing for this project.
Conflict of Interest
Shaun Goodman is supported by the Heart & Stroke Foundation of Ontario/University of Toronto Polo Chair and receives research grant support and speaker/consulting honoraria from AstraZeneca, Bayer, Bristol-Myers Squibb, Daiichi-Sankyo, Eli Lilly, and Sanofi. Lars Wallentin received research grants, consulting fee, lecture fee, and travel support from AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, and Pfizer; institutional research grant, consulting fee, lecture fee, travel support, and honoraria from GlaxoSmithKline; institutional research grant from Roche Diagnostics, Merck & Co; and consulting fees from Abbott. Axel Åkerblom received consulting, lecture fees, and institutional research grant from AstraZeneca. Niclas Eriksson received institutional research grant from AstraZeneca. Robert F. Storey has received research grants and personal fees from AstraZeneca, GlyCardial Diagnostics and Thromboserin, and consulting/lecture fees from Amgen, Bayer, Bristol Myers Squibb/Pfizer, Haemonetics and Portola. Richard Weinshilboum and Liewei Wang are cofounders of and stockholders in OneOme LLC, a pharmacogenomic decision support company. John L. Black and the Mayo Clinic have licensed intellectual property to AssureX Health (Myriad) and received royalties. He is also a co-founder of OneOme LLC and, with Mayo Clinic, has intellectual property to the company and owns stock in the company. The remaining 13 authors, Joel A. Morales-Rosado, Kashish Goel, Lingxin Zhang, Saurabh Baheti, Stefan James, Gregory D. Jenkins, Suzette J. Bielinski, Hugues Sicotte, Stephen Johnson, Veronique L. Roger, Eric W. Klee, Charanjit S. Rihal, and Naveen L. Pereira, do not have a conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM.1
(DOCX 90 kb)
Rights and permissions
About this article
Cite this article
Morales-Rosado, J.A., Goel, K., Zhang, L. et al. Next-Generation Sequencing of CYP2C19 in Stent Thrombosis: Implications for Clopidogrel Pharmacogenomics. Cardiovasc Drugs Ther 35, 549–559 (2021). https://doi.org/10.1007/s10557-020-06988-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10557-020-06988-w