Abstract
Purpose
Previous studies indicate that breast cancer molecular subtypes differ with respect to their dependency on autophagy, but our knowledge of the differential expression and prognostic significance of autophagy-related biomarkers in breast cancer is limited.
Methods
Immunohistochemistry (IHC) was performed on tissue microarrays from a large population of 3992 breast cancer patients divided into training and validation cohorts. Consensus staining scores were used to evaluate the expression levels of autophagy proteins LC3B, ATG4B, and GABARAP and determine the associations with clinicopathological variables and molecular biomarkers. Survival analyses were performed using the Kaplan–Meier function and Cox proportional hazards regression models.
Results
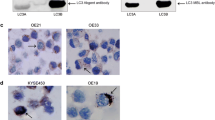
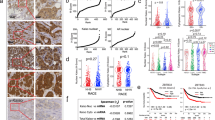
We found subtype-specific expression differences for ATG4B, with its expression lowest in basal-like breast cancer and highest in Luminal A, but there were no significant associations with patient prognosis. LC3B and GABARAP levels were highest in basal-like breast cancers, and high levels were associated with worse outcomes across all subtypes (DSS; GABARAP: HR 1.43, LC3B puncta: HR 1.43). High ATG4B levels were associated with ER, PR, and BCL2 positivity, while high LC3B and GABARAP levels were associated with ER, PR, and BCL2 negativity, as well as EGFR, HER2, HER3, CA-IX, PD-L1 positivity, and high Ki67 index (p < 0.05 for all associations). Exploratory multi-marker analysis indicated that the combination of ATG4B and GABARAP with LC3B could be useful for further stratifying patient outcomes.
Conclusions
ATG4B levels varied across breast cancer subtypes but did not show prognostic significance. High LC3B expression and high GABARAP expression were both associated with poor prognosis and with clinicopathological characteristics of aggressive disease phenotypes in all breast cancer subtypes.






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AIC:
-
Akaike information criterion
- ATG:
-
Autophagy-related
- ATG4B:
-
Autophagy-related 4B
- BCL2:
-
B-cell lymphoma 2
- CA-IX:
-
Carbonic anhydrase 9
- DSS:
-
Disease-specific survival
- EGFR:
-
Epidermal growth factor receptor
- ER:
-
Estrogen receptor
- FDA:
-
Food and Drug Administration
- GABARAP:
-
GABA type A receptor-associated protein
- GATE-16:
-
Golgi-associated ATPase enhancer of 16 kDa
- GABARAPL2:
-
GABA type A receptor-associated protein like 2
- HER2:
-
Human epidermal growth factor receptor 2
- HER3:
-
Human epidermal growth factor receptor 3
- HR:
-
Hazard ratio
- IHC:
-
Immunohistochemistry
- LC3B:
-
Microtubule associated protein 1 light chain 3 beta
- OS:
-
Overall survival
- PD-L1:
-
Programmed death-ligand 1
- PE:
-
Phosphatidylethanolamine
- PR:
-
Progesterone receptor
- RFS:
-
Relapse-free survival
- TMA:
-
Tissue microarray
- TNBC:
-
Triple-negative breast cancer
- TNP:
-
Triple-negative phenotype
References
Galluzzi L, Pietrocola F, Bravo-San Pedro JM et al (2015) Autophagy in malignant transformation and cancer progression. EMBO J 34:856–880
Amaravadi R, Kimmelman AC, White E (2016) Recent insights into the function of autophagy in cancer. Genes Dev 30:1913–1930
Levy JMM, Towers CG, Thorburn A (2017) Targeting autophagy in cancer. Nat Rev Cancer 17:528–542
Dolgin E (2019) Anticancer autophagy inhibitors attract “resurgent” interest. Nat Rev Drug Discov 18:408–410
Ravikumar B, Sarkar S, Davies JE et al (2010) Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90:1383–1435
Yang Z, Klionsky DJ (2010) Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22:124–131
Itakura E, Mizushima N (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6:764–776
Feng Y, He D, Yao Z et al (2014) The machinery of macroautophagy. Cell Res 24:24–41
Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132
Nguyen TN, Padman BS, Usher J et al (2016) Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 251:857
Weidberg H, Shvets E, Shpilka T et al (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 29:1792–1802
Shpilka T, Weidberg H, Pietrokovski S et al (2011) Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 12:226
Schaaf MBE, Keulers TG, Vooijs MA et al (2016) LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J 30:3961–3978
Tanida I, Sou Y, Ezaki J et al (2004) HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem 279:36268–36276
Satoo K, Noda NN, Kumeta H et al (2009) The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 28:1341–1350
Fujita N, Hayashi-Nishino M, Fukumoto H et al (2008) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19:4651–4659
Li M, Hou Y, Wang J et al (2011) Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem 286:7327–7338
Kauffman KJ, Yu S, Jin J et al (2018) Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy 14:992–1010
Akin D, Wang SK, Habibzadegah-Tari P et al (2014) A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 10:2021–2035
Kurdi A, Cleenewerck M, Vangestel C et al (2017) ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem Pharmacol 138:150–162
Rothe K, Lin H, Lin KBL et al (2014) The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood 123:3622–3634
Tran E, Chow A, Goda T et al (2013) Context-dependent role of ATG4B as target for autophagy inhibition in prostate cancer therapy. Biochem Biophys Res Commun 441:726–731
Bosc D, Vezenkov L, Bortnik S et al (2018) A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B. Sci Rep 8:11653
Fu Y, Hong L, Xu J et al (2018) Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy 15:1–17
Liu P-F, Tsai K-L, Hsu C-J et al (2018) Drug repurposing screening identifies tioconazole as an ATG4 inhibitor that suppresses autophagy and sensitizes cancer cells to chemotherapy. Theranostics 8:830–845
Bortnik S, Choutka C, Horlings HM et al (2016) Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 7:66970–66988
Liu P-F, Leung C-M, Chang Y-H et al (2014) ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy 10:1454–1465
Liu P-F, Hsu C-J, Tsai W-L et al (2017) Ablation of ATG4B suppressed autophagy and activated AMPK for cell cycle arrest in cancer cells. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol 44:728–740
Bortnik S, Gorski SM (2017) Clinical applications of autophagy proteins in cancer: from potential targets to biomarkers. Int J Mol Sci 18:1496
Lazova R, Camp RL, Klump V et al (2012) Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res Off J Am Assoc Cancer Res 18:370–379
Ladoire S, Chaba K, Martins I et al (2012) Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 8:1175–1184
Lefort S, Joffre C, Kieffer Y et al (2014) Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy 10:2122–2142
Ladoire S, Penault-Llorca F, Senovilla L et al (2015) Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy 11:1878–1890
Holt SV, Wyspianska B, Randall KJ et al (2011) The development of an immunohistochemical method to detect the autophagy-associated protein LC3-II in human tumor xenografts. Toxicol Pathol 39:516–523
Martinet W, Schrijvers DM, Timmermans J-P et al (2013) Immunohistochemical analysis of macroautophagy. Autophagy 9:386–402
Bertucci F, Finetti P, Cervera N et al (2008) How basal are triple-negative breast cancers? Int J Cancer 123:236–240
Perou CM (2011) Molecular stratification of triple-negative breast cancers. Oncologist 16(Suppl 1):61–70
Maycotte P, Gearheart CM, Barnard R et al (2014) STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res 74:2579–2590
Maycotte P, Thorburn A (2014) Targeting autophagy in breast cancer. World J Clin Oncol 5:224–240
Hayes DF, Ethier S, Lippman ME (2006) New guidelines for reporting of tumor marker studies in breast cancer research and treatment: REMARK. Breast Cancer Res Treat 100:237–238
Zhao H, Yang M, Zhao J et al (2013) High expression of LC3B is associated with progression and poor outcome in triple-negative breast cancer. Med Oncol Northwood Lond Engl 30:475
Chen S, Jiang Y-Z, Huang L et al (2013) The residual tumor autophagy marker LC3B serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Clin Cancer Res Off J Am Assoc Cancer Res 19:6853–6862
Klionsky DJ, Abdelmohsen K, Abe A et al (2016) Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222
Chittaranjan S, Bortnik S, Gorski SM (2015) Monitoring autophagic flux by using lysosomal inhibitors and western blotting of endogenous MAP1LC3B. Cold Spring Harb Protoc 2015:743–750
He H, Yang Y, Xiang Z et al (2016) A sensitive IHC method for monitoring autophagy-specific markers in human tumor xenografts. J Biomark. https://doi.org/10.1155/2016/1274603
Yu Z-Q, Ni T, Hong B et al (2012) Dual roles of Atg8−PE deconjugation by Atg4 in autophagy. Autophagy 8:883–892
Yang Z, Wilkie-Grantham RP, Yanagi T et al (2015) ATG4B (autophagin-1) phosphorylation modulates autophagy. J Biol Chem 290:26549–26561
Huang T, Kim CK, Alvarez AA et al (2017) MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell 32(840–855):e8
Liu P-F, Chen H-C, Cheng J-S et al (2019) Association of ATG4B and phosphorylated ATG4B proteins with tumorigenesis and prognosis in oral squamous cell carcinoma. Cancers 11:1854
Cheang MCU, Voduc D, Bajdik C et al (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res Off J Am Assoc Cancer Res 14:1368–1376
Cheang MCU, Chia SK, Voduc D et al (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101:736–750
Liu S, Chia SK, Mehl E et al (2010) Progesterone receptor is a significant factor associated with clinical outcomes and effect of adjuvant tamoxifen therapy in breast cancer patients. Breast Cancer Res Treat 119:53–61
Cheang MCU, Treaba DO, Speers CH et al (2006) Immunohistochemical detection using the new rabbit monoclonal antibody SP1 of estrogen receptor in breast cancer is superior to mouse monoclonal antibody 1D5 in predicting survival. J Clin Oncol Off J Am Soc Clin Oncol 24:5637–5644
Jensen KC, Turbin DA, Leung S et al (2008) New cutpoints to identify increased HER2 copy number: analysis of a large, population-based cohort with long-term follow-up. Breast Cancer Res Treat 112:453–459
Dawson S-J, Makretsov N, Blows FM et al (2010) BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer 103:668–675
Lou Y, McDonald PC, Oloumi A et al (2011) Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res 71:3364–3376
Chiu CG, Masoudi H, Leung S et al (2010) HER-3 overexpression is prognostic of reduced breast cancer survival: a study of 4046 patients. Ann Surg 251:1107–1116
Yerushalmi R, Gelmon KA, Leung S et al (2012) Insulin-like growth factor receptor (IGF-1R) in breast cancer subtypes. Breast Cancer Res Treat 132:131–142
Chang S-J, Ou-Yang F, Tu H-P et al (2016) Decreased expression of autophagy protein LC3 and stemness (CD44+/CD24−/low) indicate poor prognosis in triple-negative breast cancer. Hum Pathol 48:48–55
Goossens N, Nakagawa S, Sun X et al (2015) Cancer biomarker discovery and validation. Transl Cancer Res 4:256–269
Voduc D, Cheang M, Nielsen T (2008) GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res 17:365–373
Burugu S, Gao D, Leung S et al (2017) LAG-3+ tumor infiltrating lymphocytes in breast cancer: clinical correlates and association with PD-1/PD-L1+ tumors. Ann Oncol Off J Eur Soc Med Oncol 28:2977–2984
Chia S, Norris B, Speers C et al (2008) Human epidermal growth factor receptor 2 overexpression as a prognostic factor in a large tissue microarray series of node-negative breast cancers. J Clin Oncol Off J Am Soc Clin Oncol 26:5697–5704
McHugh ML (2012) Interrater reliability: the kappa statistic. Biochem Med 22:276–282
Acknowledgements
We thank Christine Chow, Angela Cheng, and Rebecca Wu for technical support and members of the Gorski laboratory for helpful comments and discussions.
Funding
This study was funded by CIHR in partnership with Avon Foundation for Women-Canada grant OBC127216 and CIHR PJT-159536 Grant to SMG. SBo was supported by a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral Award.
Author information
Authors and Affiliations
Contributions
SBo, KAG, and SMG conceived the study; SBo, BTC, and FD scored slides with assistance of JM and KG; SL, KA, and SBu performed statistical analyses; JX created KO lines and performed GABARAP antibody validation; SY and TN supervised scoring; KAG, TON, and SMG supervised and directed the study; SBo, BTC, and SL wrote the manuscript; all authors read, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
TON reports a licensing agreement with Veracyte, Inc. regarding the PAM50 intrinsic subtype classifier, not used in this study. The other authors declare that they have no conflict of interest.
Ethical approval
This study and our access to the de-identified human data were approved by the Research Ethics Board of the University of British Columbia, British Columbia Cancer Agency and Simon Fraser University. This article does not contain any studies with animals performed by any of the authors.
Informed consent
No informed consent was needed as anonymized archival specimens were used in this study, according to the Canadian Tri-Council Policy Statement for ethical research involving human subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bortnik, S., Tessier-Cloutier, B., Leung, S. et al. Differential expression and prognostic relevance of autophagy-related markers ATG4B, GABARAP, and LC3B in breast cancer. Breast Cancer Res Treat 183, 525–547 (2020). https://doi.org/10.1007/s10549-020-05795-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05795-z