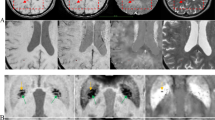
Abstract
Cerebral microbleeds, which are small focal hemorrhages in the brain that are prevalent in many diseases, are gaining increasing attention due to their potential as surrogate markers of disease burden, clinical outcomes, and delayed effects of therapy. Manual detection is laborious and automatic detection and labeling of these lesions is challenging using traditional algorithms. Inspired by recent successes of deep convolutional neural networks in computer vision, we developed a 3D deep residual network that can distinguish true microbleeds from false positive mimics of a previously developed technique based on traditional algorithms. A dataset of 73 patients with radiation-induced cerebral microbleeds scanned at 7 T with susceptibility-weighted imaging was used to train and evaluate our model. With the resulting network, we maintained 95% of the true microbleeds in 12 test patients and the average number of false positives was reduced by 89%, achieving a detection precision of 71.9%, higher than existing published methods. The likelihood score predicted by the network was also evaluated by comparing to a neuroradiologist’s rating, and good correlation was observed.






Similar content being viewed by others
Change history
08 February 2019
This paper was published inadvertently as open access. It has been corrected online.
References
Linn J, Halpin A, Demaerel P, Ruhland J, Giese AD, Dichgans M, van Buchem MA, Bruckmann H, Greenberg SM: Prevalence of superficial siderosis in patients with cerebral amyloid angiopathy. Neurology 74(17):1346–1350, 2010
Kato H, Izumiyama M, Izumiyama K, Takahashi A, Itoyama Y: Silent cerebral microbleeds on T2*-weighted MRI: correlation with stroke subtype, stroke recurrence, and leukoaraiosis. Stroke 33(6):1536–1540, 2002
Hanyu H, Tanaka Y, Shimizu S, Takasaki M, Abe K: Cerebral microbleeds in Alzheimer’s disease. J Neurol 250(12):1496–1497, 2003
Kinnunen KM, Greenwood R, Powell JH, Leech R, Hawkins PC, Bonnelle V, Patel MC, Counsell SJ, Sharp DJ: White matter damage and cognitive impairment after traumatic brain injury. Brain 134(Pt 2):449–463, 2011
Lupo JM, Chuang CF, Chang SM, Barani IJ, Jimenez B, Hess CP, Nelson SJ. 7-Tesla susceptibilityweighted imaging to assess the effects of radiotherapy on normal-appearing brain in patients with glioma. Int J Radiat Oncol Biol Phys 82(3):e493–500, 2012
Charidimou A, Krishnan A, Werring DJ, Jäger HR: Cerebral microbleeds: a guide to detection and clinical relevance in different disease settings. Neuroradiology 55(6):655–674, 2013
Wahl M, Anwar M, Hess C, Chang SM, Lupo JM: Relationship between radiation dose and microbleed formation in patients with malignant glioma. Int J Radiat Oncol Biol Phys 96(2S):E68, 2016
Kuijf HJ, de Bresser J, Geerlings MI, Conijn MMA, Viergever MA, Biessels GJ, Vincken KL: Efficient detection of cerebral microbleeds on 7.0 T MR images using the radial symmetry transform. Neuroimage 59(3):2266–2273, 2012
van den Heuvel TLA, Ghafoorian M, van der Eerden AW, Goraj BM, Andriessen TMJC, ter Haar Romeny BM, Platel B: Computer aided detection of brain micro-bleeds in traumatic brain injury. In: Medical Imaging 2015: Computer-Aided Diagnosis, Vol. 9414, 2015, p. 94142F
Barnes SRS, Haacke EM, Ayaz M, Boikov AS, Kirsch W, Kido D: Semiautomated detection of cerebral microbleeds in magnetic resonance images. Magn Reson Imaging 29(6):844–852, 2011
Qi D, Chen H, Lequan Y, Zhao L, Qin J, Wang D, Mok VC, Shi L, Heng P-A: Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 35(5):1182–1195, 2016
Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90, 2017
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L: ImageNet large scale visual recognition challenge. Int J Comput Vis 115(3):211–252, 2015
He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 770–778
Huang G, Liu Z, van der Maaten L, Weinberger KQ: Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 2261–2269
Ioffe S, Szegedy C: Batch normalization: accelerating deep network training by reducing internal covariate shift, arXiv preprint arXiv:1502.03167, 2015
Bian W, Hess CP, Chang SM, Nelson SJ, Lupo JM: Computer-aided detection of radiation-induced cerebral microbleeds on susceptibility-weighted MR images. Neuroimage Clin 2:282–290, 2013
Bian W, Banerjee S, Kelly DAC, Hess CP, Larson PEZ, Chang SM, Nelson SJ, Lupo JM: Simultaneous imaging of radiation-induced cerebral microbleeds, arteries and veins, using a multiple gradient echo sequence at 7 Tesla. J Magn Reson Imaging 42(2):269–279, 2015
Lupo JM, Banerjee S, Hammond KE, Kelley DAC, Xu D, Chang SM, Vigneron DB, Majumdar S, Nelson SJ: GRAPPA-based susceptibility-weighted imaging of normal volunteers and patients with brain tumor at 7 T. Magn Reson Imaging 27(4):480–488, 2009
Smith SM: Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155, 2002
Dahl GE, Sainath TN, Hinton GE: Improving deep neural networks for LVCSR using rectified linear units and dropout, Acoustics, Speech and Signal Processing (ICASSP), IEEE International Conference on, 2013, p 8609
F. Chollet and Others, Keras, 2015.
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S: Tensorflow: Large-scale machine learning on heterogeneous distributed systems, arXiv preprint arXiv:1603.04467, 2016.
Kingma DP, Ba J: Adam: a method for stochastic optimization, arXiv preprint arXiv:1412.6980, 2014
Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556, 2014
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A: Going deeper with convolutions. In 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015, pp 1–9
Ren S, He K, Girshick R, Sun J: Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 39(6):1137–1149, 2017
Funding
This work was supported by the National Institute for Child Health and Human Development of the National Institutes of Health grant R01HD079568 and GE Healthcare.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest with regard to the content of this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: with the author's decision to step back from Open Choice, the copyright of the article changed to © Society for Imaging Informatics in Medicine 2018 and the article is forthwith distributed under the terms of copyright.
Rights and permissions
About this article
Cite this article
Chen, Y., Villanueva-Meyer, J.E., Morrison, M.A. et al. Toward Automatic Detection of Radiation-Induced Cerebral Microbleeds Using a 3D Deep Residual Network. J Digit Imaging 32, 766–772 (2019). https://doi.org/10.1007/s10278-018-0146-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-018-0146-z