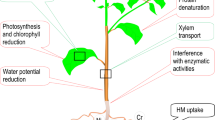
Abstract
High explosives such as hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), and 2,4,6-trinitrotoluene (TNT) are important contaminants in the environment and phytoremediation has been viewed as a cost-effective abatement. There remains, however, an insufficient knowledge-base about how plants respond to explosives, especially in the steady state. Microarray analysis was conducted on Arabidopsis thaliana that were grown in Murashige and Skoog media containing steady-state levels of 0.5 mM RDX or 2.0 μM TNT to study the effect of these compounds on its transcriptional profile. Our results for both RDX and TNT were consistent with the existing theory for xenobiotic metabolism in plants. Among the genes that were differentially expressed included oxidoreductases, cytochrome P450s, transferases, transporters, and several unknown expressed proteins. We discuss the potential role of upregulated genes in plant metabolism, phytoremediation, and phytosensing. Phytosensing, the detection of field contamination using plants, is an end goal of this project.


Similar content being viewed by others
References
Abercrombie JM, Halfhill MD, Ranjan P, Rao MR, Saxton AM, Yuan JS, Stewart CN Jr (2008) Transcriptional responses of Arabidopsis thaliana plants to As (V) stress. BMC Plant Biol 8:87
Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, Simmons MP, Bedinger P, Goff SA, Ringli C, Keller B (2003) Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol 131:1313–1326
Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92:8675–8679
Best EPH, Zappi ME, Fredrickson HL, Sprecher SL, Larson SL, Ochman M (1997) Screening of aquatic and wetland plant species for phytoremediation of explosives-contaminated groundwater from the Iowa Army Ammunition Plant. Ann N Y Acad Sci 829:179–194
Best EPH, Sprecher SL, Larson SL, Fredrickson HL, Bader DF (1999) Environmental behavior of explosives in groundwater from the Milan Army Ammunition Plant in aquatic and wetland plant treatments. Uptake and fate of TNT and RDX in plants. Chemosphere 39:2057–2072
Best EPH, Miller JL, Larson SL (2001) Tolerance towards explosives, and explosives removal from groundwater in treatment wetland mesocosms. Water Sci Technol 44:515–521
Best EPH, Kvesitadze G, Khatisashvili G, Sadunishvili T (2005) Plant processes important for the transformation and degradation of explosives contaminants. Z Naturforsch C J Biosci 60:340–348
Best EPH, Geter KN, Tatem HE, Lane BK (2006) Effects, transfer, and fate of RDX from aged soil in plants and worms. Chemosphere 62:616–625
Bhadra R, Spanggord RJ, Wayment DG, Hughes JB, Shanks JV (1999a) Characterization of oxidation products of TNT metabolism in aquatic phytoremediation systems of Myriophyllum aquaticum. Environ Sci Technol 33:3354–3361
Bhadra R, Wayment DG, Hughes JB, Shanks JV (1999b) Confirmation of conjugation processes during TNT metabolism by axenic plant roots. Environ Sci Technol 33:446–452
Bhadra R, Wayment DG, Williams RK, Barman SN, Stone MB, Hughes JB, Shanks JV (2001) Studies on plant-mediated fate of the explosives RDX and HMX. Chemosphere 44:1259–1264
Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92
Burken JG, Shanks JV, Thompson PL (2000) Phytoremediation and plant metabolism of explosives and nitroaromatic compounds. In: Spain JC, Hughes JB, Knackmuss H (eds) Biodegradation of nitroaromatic compounds and explosives. CRC, Boca Raton, FL, pp 239–276
Campbell EJ, Schenk PM, Kazan K, Penninckx IA, Anderson JP, Maclean DJ, Cammue BP, Ebert PR, Manners JM (2003) Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol 133:1272–1284
Coleman J, Blake-Kalff M, Davies E (1997) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2:144–151
Cunningham SD, Berti WR, Huang JWW (1995) Phytoremediation of contaminated soils. Trends Biotechnol 13:393–397
Eapen S, Singh S, D'Souza SF (2007) Advances in development of transgenic plants for remediation of xenobiotic pollutants. Biotechnol Adv 25:442–451
Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JFD (2003) SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2, 4, 6-trinitrotoluene. Plant Physiol 133:1397–1406
Ekman DR, Wolfe NL, Dean JFD (2005) Gene expression changes in Arabidopsis thaliana seedling roots exposed to the munition hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine. Environ Sci Technol 39:6313–6320
Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53:275–297
French CE, Rosser SJ, Davies GJ, Nicklin S, Bruce NC (1999) Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol 17:491–494
Gandia-Herrero F, Lorenz A, Larson T, Graham IA, Bowles DJ, Rylott EL, Bruce NC (2008) Detoxification of the explosive 2, 4, 6-trinitrotoluene in Arabidopsis: discovery of bifunctional O- and C-glucosyltransferases. Plant J 56:963–974
Halasz A, Groom C, Zhou E, Paquet L, Beaulieu C, Deschamps S, Corriveau A, Thiboutot S, Ampleman G, Dubois C, Hawari J (2002) Detection of explosives and their degradation products in soil environments. J Chromatogr A 963:411–418
Hannink N, Rosser SJ, French CE, Basran A, Murray JAH, Nicklin S, Bruce NC (2001) Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase. Nat Biotechnol 19:1168–1172
Hannink NK, Rosser SJ, Bruce NC (2002) Phytoremediation of explosives. Crit Rev Plant Sci 21:511–538
Hannink NK, Subramanian M, Rosser SJ, Basran A, Murray JAH, Shanks JV, Bruce NC (2007) Enhanced transformation of TNT by tobacco plants expressing a bacterial nitroreductase. Int J Phytoremediation 9:385–401
Harvey SD, Fellows RJ, Cataldo DA, Bean RM (1990) Analysis of 2, 4, 6-trinitrotoluene and its transformation products in soils and plant-tissues by high-performance liquid-chromatography. J Chromatogr 518:361–374
Harvey SD, Fellows RJ, Cataldo DA, Bean RM (1991) Fate of the explosive hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) in soil and bioaccumulation in bush bean hydroponic plants. Environ Toxicol Chem 10:845–855
Hawari J, Beaudet S, Halasz A, Thiboutot S, Ampleman G (2000) Microbial degradation of explosives: biotransformation versus mineralization. Appl Microbiol Biotechnol 54:605–618
Hughes JB, Shanks J, Vanderford M, Lauritzen J, Bhadra R (1997) Transformation of TNT by aquatic plants and plant tissue cultures. Environ Sci Technol 31:266–271
Ishikawa T (1992) The ATP-dependent glutathione S-conjugate export pump. Trends Biochem Sci 17:463–469
Ishikawa T, Li ZS, Lu YP, Rea PA (1997) The GS-X pump in plant, yeast, and animal cells: structure, function, and gene expression. Biosci Rep 17:189–207
Just CL, Schnoor JL (2004) Phytophotolysis of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) in leaves of reed canary grass. Environ Sci Technol 38:290–295
Kobe B, Deisenhofer J (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19:415–421
Kulkarni AP (2001) Lipoxygenase—a versatile biocatalyst for biotransformation of endobiotics and xenobiotics. Cell Mol Life Sci 58:1805–1825
Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J (2005) Independence and reproducibility across microarray platforms. Nat Meth 2:337–344
Larson SL, Jones RP, Escalon L, Parker D (1999) Classification of explosives transformation products in plant tissue. Environ Toxicol Chem 18:1270–1276
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Mentewab A, Cardoza V, Stewart CN (2005) Genomic analysis of the response of Arabidopsis thaliana to trinitrotoluene as revealed by cDNA microarrays. Plant Sci 168:1409–1424
Mezzari MP, Walters K, Jelinkova M, Shih MC, Just CL, Schnoor JL (2005) Gene expression and microscopic analysis of Arabidopsis exposed to chloroacetanilide herbicides and explosive compounds. A phytoremediation approach. Plant Physiol 138:858–869
Nishino SF, Spain JC, He Z (2000) Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application. In: Spain JC, Hughes JB, Knackmuss H (eds) Biodegradation of nitroaromatic compounds and explosives. CRC, Boca Raton, FL, pp 7–61
Petersen D, Chandramouli GVR, Geoghegan J, Hilburn J, Paarlberg J, Kim C, Munroe D, Gangi L, Han J, Puri R, Staudt L, Weinstein J, Barrett JC, Green J, Kawasaki E (2005) Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics 6:63
Peterson MM, Horst GL, Shea PJ, Comfort SD (1998) Germination and seedling development of switchgrass and smooth bromegrass exposed to 2, 4, 6-trinitrotoluene. Environ Pollut 99:53–59
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Purugganan MM, Braam J, Fry SC (1997) The Arabidopsis TCH4 xyloglucan endotransglycosylase (substrate specificity, pH optimum, and cold tolerance). Plant Physiol 115:181–190
Pylatuik JD, Fobert PR (2005) Comparison of transcript profiling on Arabidopsis microarray platform technologies. Plant Mol Biol 58:609–624
Rea PA, Li ZS, Lu YP, Drozdowicz YM, Martinoia E (1998) From vacuolar GS-X pumps to multispecific ABC transporters. Annu Rev Plant Physiol Plant Mol Biol 49:727–760
Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller LA, Mundodi S, Reiser L, Tacklind J, Weems DC, Wu Y, Xu I, Yoo D, Yoon J, Zhang P (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31:224–228
Rogojina AT, Orr WE, Song BK, Geisert EE Jr (2003) Comparing the use of Affymetrix to spotted oligonucleotide microarrays using two retinal pigment epithelium cell lines. Mol Vis 9:482–496
Rosenblatt DH (1980) Toxicology of explosives and propellants. In: Kaye SM (ed) Encyclopedia of explosives and related items. US Army Armament Research Development Committee, Dover, NJ
Rosenblatt DH, Burrows SP, Mitchell WR, Parmer DL (1991) Organic explosives and related compounds. In: Hutzinger O (ed) The handbook of environmental chemistry. Springer, New York
Rylott EL, Bruce NC (2009) Plants disarm soil: engineering plants for the phytoremediation of explosives. Trends Biotechnol 27:73–81
Rylott EL, Jackson RG, Edwards J, Womack GL, Seth-Smith HM, Rathbone DA, Strand SE, Bruce NC (2006) An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat Biotechnol 24:216–219
Sandermann H Jr (1992) Plant metabolism of xenobiotics. Trends Biochem Sci 17:82–84
Schaffner A, Messner B, Langebartels C, Sandermann H (2002) Genes and enzymes for in-planta phytoremediation of air, water and soil. Acta Biotechnol 22:141–151
Shi LM, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo YL, Sun YMA, Willey JC, Setterquist RA, Fischer GM et al (2006) The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 24:1151–1161
Tan PK, Downey TJ, Spitznagel EL Jr, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC (2003) Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res 31:5676–5684
Tanaka S, Brentner LB, Merchie KM, Schnoor JL, Jong MY, Van Aken B (2007) Analysis of gene expression in poplar trees (Populus deltoides x nigra, DN34) exposed to the toxic explosive hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX). Int J Phytoremediation 9:15–30
Thompson PL, Ramer LA, Schnoor JL (1999) Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine translocation in poplar trees. Environ Toxicol Chem 18:279–284
Van AB (2009) Transgenic plants for enhanced phytoremediation of toxic explosives. Curr Opin Biotechnol. doi:10.1016/j.copbio.2009.01.011
Van AB, Yoon JM, Just CL, Schnoor JL (2004) Metabolism and mineralization of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine inside poplar tissues (Populus deltoides x nigra DN-34). Environ Sci Technol 38:4572–4579
Van Dillewijn P, Caballero A, Paz JA, Gonzalez-Perez MM, Oliva JM, Ramos JL (2007) Bioremediation of 2, 4, 6-trinitrotoluene under field conditions. Environ Sci Technol 41:1378–1383
Van Dillewijn P, Couselo JL, Corredoira E, Delgado A, Wittich RM, Ballester A, Ramos JL (2008) Bioremediation of 2, 4, 6-trinitrotoluene by bacterial nitroreductase expressing transgenic aspen. Environ Sci Technol 42:7405–7410
Vila M, Lorber-Pascal S, Laurent F (2007) Fate of RDX and TNT in agronomic plants. Environ Poll 148:148–154
Acknowledgments
We would like to thank Pradeep Chimakurthy for the help on Affymetrix data analysis and all the members of Stewart lab for their support. Also, we would like to thank the reviewers for their constructive comments and suggestions. We are grateful for the funding by DIA-AFMIC and the Tennessee Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Significant genes upregulated (≥1.5 folds and ≤10% FDR) in response to RDX (Arabidopsis seedlings exposed to 0.5 mM of RDX for 8–9 days) in the two-color microarrays along with their linear fold change (FC), false discovery rate (FDR), and the p value associated with the genes (XLS 25 kb)
Table S2
Significant genes downregulated (≤1.5 folds and ≤10% FDR) in response to RDX (Arabidopsis seedlings exposed to 0.5 mM of RDX for 8–9 days) in the two-color microarrays along with their linear fold change (FC), false discovery rate (FDR), and the p value associated with the genes (XLS 22 kb)
Table S3
Significant genes upregulated (≥2.0 folds and ≤10% FDR) in response to RDX (Arabidopsis seedlings exposed to 0.5 mM of RDX for 8–9 days) in the Affymetrix microarrays along with their linear fold change (FC), false discovery rate (FDR), and the p value associated with the genes (XLS 24 kb)
Table S4
Significant genes downregulated (≤2.0 folds and ≤10% FDR) in response to RDX (Arabidopsis seedlings exposed to 0.5 mM of RDX for 8–9 days) in the Affymetrix microarrays along with their linear fold change (FC), false discovery rate (FDR), and the p value associated with the genes (XLS 24 kb)
Table S5
Significant genes upregulated (≥2.0 folds and ≤10% FDR) in response to TNT (Arabidopsis seedlings exposed to 2.0 μM of TNT for 8–9 days) in the Affymetrix microarrays along with their linear fold change (FC), false discovery rate (FDR), and the p value associated with the genes (XLS 25 kb)
Table S6
Significant genes downregulated (≤2.0 folds and ≤10% FDR) in response to TNT (Arabidopsis seedlings exposed to 2.0 μM of TNT for 8–9 days) in the Affymetrix microarrays along with their linear fold change (FC), false discovery rate (FDR), and the p value associated with the genes (XLS 26 kb)
Table S7
Real-time RT-PCR confirmation of selected differentially regulated genes in the RDX microarray experiments (XLS 14 kb)
Table S8
Significant genes commonly up- and downregulated between RDX Affymetrix and RDX two-color along with their linear fold change (FC), false discovery rate (FDR), and associated p values (XLS 19 kb)
Fig. S1
Arabidopsis thaliana (ecotype Columbia) grown on MS medium supplemented with different concentrations of RDX. (PDF 826 kb)
Fig. S2
Arabidopsis thaliana (ecotype Columbia) grown on MS medium supplemented with different concentrations of TNT. (PDF 789 kb)
Fig. S3
Gene bar charts for functional categorization by molecular function for genes differentially expressed in the RDX and TNT microarrays. (PDF 95 kb)
Fig. S4
Gene bar charts for functional categorization by cellular component and biological process for genes differentially expressed in the RDX two-color microarrays. (PDF 66 kb)
Fig. S5
Gene bar charts for functional categorization by cellular component and biological process for genes differentially expressed in the RDX Affymetrix microarrays. (PDF 70 kb)
Fig. S6
Gene bar charts for functional categorization by cellular component and biological process for genes differentially expressed in the TNT Affymetrix microarrays. (PDF 68 kb)
Rights and permissions
About this article
Cite this article
Rao, M.R., Halfhill, M.D., Abercrombie, L.G. et al. Phytoremediation and phytosensing of chemical contaminants, RDX and TNT: identification of the required target genes. Funct Integr Genomics 9, 537–547 (2009). https://doi.org/10.1007/s10142-009-0125-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-009-0125-z