Abstract
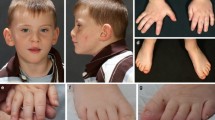
Molecular anomalies in MED13L, leading to haploinsufficiency, have been reported in patients with moderate to severe intellectual disability (ID) and distinct facial features, with or without congenital heart defects. Phenotype of the patients was referred to “MED13L haploinsufficiency syndrome.” Missense variants in MED13L were already previously described to cause the MED13L-related syndrome, but only in a limited number of patients. Here we report 36 patients with MED13L molecular anomaly, recruited through an international collaboration between centers of expertise for developmental anomalies. All patients presented with intellectual disability and severe language impairment. Hypotonia, ataxia, and recognizable facial gestalt were frequent findings, but not congenital heart defects. We identified seven de novo missense variations, in addition to protein-truncating variants and intragenic deletions. Missense variants clustered in two mutation hot-spots, i.e., exons 15–17 and 25–31. We found that patients carrying missense mutations had more frequently epilepsy and showed a more severe phenotype. This study ascertains missense variations in MED13L as a cause for MED13L-related intellectual disability and improves the clinical delineation of the condition.


Similar content being viewed by others
References
Allen BL, Taatjes DJ (2015) The mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16(3):155–166
Yin J-w, Wang G (2014) The mediator complex: a master coordinator of transcription and cell lineage development. Development 141(5):977–987
Daniels DL (2013) Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK-mediator kinase module. J Proteomics Bioinformat 01(S2)
Graham JM, Schwartz CE (2013) MED12 related disorders. Am J Med Genet A 161A(11):2734–2740
Mukhopadhyay A, Kramer JM, Merkx G, Lugtenberg D, Smeets DF, Oortveld MAW, Blokland EAW, Agrawal J, Schenck A, van Bokhoven H, Huys E, Schoenmakers EF, van Kessel AG, van Nouhuys CE, Cremers FPM (2010) CDK19 is disrupted in a female patient with bilateral congenital retinal folds, microcephaly and mild mental retardation. Hum Genet 128(3):281–291
Adegbola A, Musante L, Callewaert B, Maciel P, Hu H, Isidor B, Picker-Minh S, le Caignec C, Delle Chiaie B, Vanakker O, Menten B, Dheedene A, Bockaert N, Roelens F, Decaestecker K, Silva J, Soares G, Lopes F, Najmabadi H, Kahrizi K, Cox GF, Angus SP, Staropoli JF, Fischer U, Suckow V, Bartsch O, Chess A, Ropers HH, Wienker TF, Hübner C, Kaindl AM, Kalscheuer VM (2015) Redefining the MED13L syndrome. Eur J Hum Genet 23(10):1308–1317
Asadollahi R, Oneda B, Sheth F, Azzarello-Burri S, Baldinger R, Joset P, Latal B, Knirsch W, Desai S, Baumer A, Houge G, Andrieux J, Rauch A (2013) Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet 21(10):1100–1104
Asadollahi R et al. Genotype-phenotype evaluation of MED13L defects in the light of a novel truncating and a recurrent missense mutation. Eur J Med Genet 60(9):451-464
Muncke N, Jung C, Rüdiger H, Ulmer H, Roeth R, Hubert A, Goldmuntz E, Driscoll D, Goodship J, Schön K, Rappold G (2003) Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries). Circulation 108(23):2843–2850
van Haelst MM, Monroe GR, Duran K, van Binsbergen E, Breur JM, Giltay JC, van Haaften G (2015) Further confirmation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet 23(1):135–138
2017/06/17/09:38:34
Cafiero C, Marangi G, Orteschi D, Ali M, Asaro A, Ponzi E, Moncada A, Ricciardi S, Murdolo M, Mancano G, Contaldo I, Leuzzi V, Battaglia D, Mercuri E, Slavotinek AM, Zollino M (2015) Novel de novo heterozygous loss-of-function variants in MED13L and further delineation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet 23(11):1499–1504
Caro-Llopis A, Rosello M, Orellana C, Oltra S, Monfort S, Mayo S, Martinez F (2016) De novo mutations in genes of mediator complex causing syndromic intellectual disability: mediatorpathy or transcriptomopathy? Pediatr Res 80:809–815
Deciphering Developmental Disorders, S (2017) Prevalence and architecture of de novo mutations in developmental disorders. Nature 542(7642):433–438
Gordon CT, Chopra M, Oufadem M, Alibeu O, Bras M, Boddaert N, Bole-Feysot C, Nitschké P, Abadie V, Lyonnet S, Amiel J (2018) MED13L loss-of-function variants in two patients with syndromic Pierre Robin sequence. Am J Med Genet A 176(1):181–186
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB, Mardis ER, Wilson RK, Schatz MC, McCombie WR, Wigler M (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74(2):285–299
Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BBA, Kleefstra T, Brunner HG, Vissers LELM, Veltman JA (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature 511(7509):344–347
Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, Massicotte C, Ambalavanan A, Spiegelman D, Diallo O, Henrion E, Dionne-Laporte A, Fougerat A, Pshezhetsky AV, Venkateswaran S, Rouleau GA, Michaud JL (2014) De novo mutations in moderate or severe intellectual disability. PLoS Genet 10(10):e1004772
Redin C, Gérard B, Lauer J, Herenger Y, Muller J, Quartier A, Masurel-Paulet A, Willems M, Lesca G, el-Chehadeh S, le Gras S, Vicaire S, Philipps M, Dumas M, Geoffroy V, Feger C, Haumesser N, Alembik Y, Barth M, Bonneau D, Colin E, Dollfus H, Doray B, Delrue MA, Drouin-Garraud V, Flori E, Fradin M, Francannet C, Goldenberg A, Lumbroso S, Mathieu-Dramard M, Martin-Coignard D, Lacombe D, Morin G, Polge A, Sukno S, Thauvin-Robinet C, Thevenon J, Doco-Fenzy M, Genevieve D, Sarda P, Edery P, Isidor B, Jost B, Olivier-Faivre L, Mandel JL, Piton A (2014) Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet 51(11):724–736
Utami KH, Winata CL, Hillmer AM, Aksoy I, Long HT, Liany H, Chew EG, Mathavan S, Tay SK, Korzh V, Sarda P, Davila S, Cacheux V (2014) Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum Mutat 35(11):1311–1320
Wang T, Guo H, Xiong B, Stessman HAF, Wu H, Coe BP, Turner TN, Liu Y, Zhao W, Hoekzema K, Vives L, Xia L, Tang M, Ou J, Chen B, Shen Y, Xun G, Long M, Lin J, Kronenberg ZN, Peng Y, Bai T, Li H, Ke X, Hu Z, Zhao J, Zou X, Xia K, Eichler EE (2016) De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun 7:13316
Popp B, Ekici AB, Thiel CT, Hoyer J, Wiesener A, Kraus C, Reis A, Zweier C (2017) Exome Pool-Seq in neurodevelopmental disorders. Eur J Hum Genet 25(12):1364–1376
Deciphering Developmental Disorders, S (2015) Large-scale discovery of novel genetic causes of developmental disorders. Nature 519(7542):223–228
Lehalle D, Mosca-Boidron AL, Begtrup A, Boute-Benejean O, Charles P, Cho MT, Clarkson A, Devinsky O, Duffourd Y, Duplomb-Jego L, Gérard B, Jacquette A, Kuentz P, Masurel-Paulet A, McDougall C, Moutton S, Olivié H, Park SM, Rauch A, Revencu N, Rivière JB, Rubin K, Simonic I, Shears DJ, Smol T, Taylor Tavares AL, Terhal P, Thevenon J, van Gassen K, Vincent-Delorme C, Willemsen MH, Wilson GN, Zackai E, Zweier C, Callier P, Thauvin-Robinet C, Faivre L (2017) STAG1 mutations cause a novel cohesinopathy characterised by unspecific syndromic intellectual disability. J Med Genet 54:479–488
van Weerd JH, Badi I, van den Boogaard M, Stefanovic S, van de Werken HJG, Gomez-Velazquez M, Badia-Careaga C, Manzanares M, de Laat W, Barnett P, Christoffels VM (2014) A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ Res 115(4):432–441
Mullegama SV, Jensik P, Li C, Dorrani N, UCLA Clinical Genomics Center, Kantarci S, Blumberg B, Grody WW, Strom SP (2017) Coupling clinical exome sequencing with functional characterization studies to diagnose a patient with familial Mediterranean fever and MED13L haploinsufficiency syndromes. Clin Case Rep 5(6):833–840
Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ (2013) A conserved mediator-CDK8 kinase module association regulates mediator-RNA polymerase II interaction. Nat Struct Mol Biol 20(5):611–619
Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE (2013) The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with mediator. Genes Dev 27(2):151–156
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplemental Table 1
Comparison of clinical data of patients harboring MED13L protein-truncating variants and missense variants. (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Smol, T., Petit, F., Piton, A. et al. MED13L-related intellectual disability: involvement of missense variants and delineation of the phenotype. Neurogenetics 19, 93–103 (2018). https://doi.org/10.1007/s10048-018-0541-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-018-0541-0