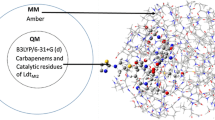
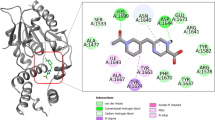
Abstract
Virtual screening is a useful in silico approach to identify potential leads against various targets. It is known that carbapenems (doripenem and faropenem) do not show any reasonable inhibitory activities against L,D-transpeptidase 5 (LdtMt5) and also an adduct of meropenem exhibited slow acylation. Since these drugs are active against L,D-transpeptidase 2 (LdtMt2), understanding the differences between these two enzymes is essential. In this study, a ligand-based virtual screening of 12,766 compounds followed by molecular dynamics (MD) simulations was applied to identify potential leads against LdtMt5. To further validate the obtained virtual screening ranking for LdtMt5, we screened the same libraries of compounds against LdtMt2 which had more experimetal and calculated binding energies reported. The observed consistency between the binding affinities of LdtMt2 validates the obtained virtual screening binding scores for LdtMt5. We subjected 37 compounds with docking scores ranging from − 7.2 to − 9.9 kcal mol−1 obtained from virtual screening for further MD analysis. A set of compounds (n = 12) from four antibiotic classes with ≤ − 30 kcal mol−1 molecular mechanics/generalized born surface area (MM-GBSA) binding free energies (ΔGbind) was characterized. A final set of that, all β-lactams (n = 4), was considered. The outcome of this study provides insight into the design of potential novel leads for LdtMt5.

Graphical abstract





Similar content being viewed by others
References
Seung KJ, Keshavjee S, Rich ML (2015) Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harbor perspectives in medicine:a017863
Billones JB, Carrillo MCO, Organo VG, Macalino SJY, Sy JBA, Emnacen IA, Clavio NAB, Concepcion GP (2016) Toward antituberculosis drugs: in silico screening of synthetic compounds against Mycobacterium tuberculosis L, D-transpeptidase 2. Drug Des Devel Ther 10:1147
Adewumi OA (2012) Treatment outcomes in patients infected with multidrug resistant tuberculosis and in patients with multidrug resistant tuberculosis coinfected with human immunodeficiency virus at Brewelskloof Hospital
Basta LAB, Ghosh A, Pan Y, Jakoncic J, Lloyd EP, Townsend CA, Lamichhane G, Bianchet MA (2015) Loss of a functionally and structurally distinct ld-transpeptidase, LdtMt5, compromises cell wall integrity in mycobacterium tuberculosis. J Biol Chem 290(42):25670–25685
Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi J-L (2008) The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L, D-transpeptidation. J Bacteriol 190(12):4360–4366
Dubée V, Triboulet S, Mainardi J-L, Ethève-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet J-E, Arthur M (2012) Inactivation of mycobacterium tuberculosis L, D-transpeptidase LdtMt1 by carbapenems and cephalosporins. Antimicrob Agents Chemother 56(8):4189–4195
Cordillot M, Dubée V, Triboulet S, Dubost L, Marie A, Hugonnet J-E, Arthur M, Mainardi J-L (2013) In vitro cross-linking of Mycobacterium tuberculosis peptidoglycan by l, d-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrob Agents Chemother 57(12):5940–5945
Lecoq L, Dubée V, Sb T, Bougault C, Hugonnet J-E, Arthur M, Simorre J-P (2013) Structure of enterococcus faecium L, D-transpeptidase acylated by ertapenem provides insight into the inactivation mechanism. ACS Chem Biol 8(6):1140–1146
Gupta R, Lavollay M, Mainardi J-L, Arthur M, Bishai WR, Lamichhane G (2010) The mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 16(4):466–469
Biarrotte-Sorin S, Hugonnet J-E, Delfosse V, Mainardi J-L, Gutmann L, Arthur M, Mayer C (2006) Crystal structure of a novel β-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol 359(3):533–538
Mainardi J-L, Fourgeaud M, Hugonnet J-E, Dubost L, Brouard J-P, Ouazzani J, Rice LB, Gutmann L, Arthur M (2005) A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. J Biol Chem 280(46):38146–38152
Mainardi J-L, Villet R, Bugg TD, Mayer C, Arthur M (2008) Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev 32(2):386–408
Tolufashe GF, Sabe VT, Ibeji CU, Ntombela T, Govender T, Maguire GE, Kruger HG, Lamichhane G, Honarparvar B (2019) Structure and function of L, D-and D, D-transpeptidase family enzymes from Mycobacterium tuberculosis. Curr Med Chem
Honarparvar B, Govender T, Maguire GE, Soliman ME, Kruger HG (2013) Integrated approach to structure-based enzymatic drug design: molecular modeling, spectroscopy, and experimental bioactivity. Chem Rev 114(1):493–537
Bradley J, Garau J, Lode H, Rolston K, Wilson S, Quinn J (1999) Carbapenems in clinical practice: a guide to their use in serious infection. Int J Antimicrob Agents 11(2):93–100
Paterson D (2000) Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs). Clin Microbiol Infect 6(9):460–463
Paterson DL Serious infections caused by enteric gram-negative bacilli--mechanisms of antibiotic resistance and implications for therapy of gram-negative sepsis in the transplanted patient. In: Seminars in respiratory infections, 2002. vol 4. pp 260–264
Paterson DL, Bonomo RA (2005) Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 18(4):657–686
Torres JA, Villegas MV, Quinn JP (2007) Current concepts in antibiotic-resistant gram-negative bacteria. Expert Rev Anti-Infect Ther 5(5):833–843
Meletis G (2016) Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis 3(1):15–21
Kattan J, Villegas M, Quinn J (2008) New developments in carbapenems. Clin Microbiol Infect 14(12):1102–1111
El-Gamal MI, Brahim I, Hisham N, Aladdin R, Mohammed H, Bahaaeldin A (2017) Recent updates of carbapenem antibiotics. Eur J Med Chem 131:185–195
Tolufashe GF, Sabe VT, Ibeji CU, Lawal MM, Govender T, Maguire GE, Lamichhane G, Kruger HG, Honarparvar B (2019) Inhibition mechanism of L, D-transpeptidase 5 in presence of the β-lactams using ONIOM method. J Mol Graph Model 87:204–210
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Schrödinger Release 2018–1: Maestro S, LLC, New York, NY, 2018
Reddy AS, Pati SP, Kumar PP, Pradeep H, Sastry GN (2007) Virtual screening in drug discovery-a computational perspective. Curr Protein Pept Sci 8(4):329–351
Roe DR, Cheatham III TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095
Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham III TE, DeBolt S, Ferguson D, Seibel G, Kollman P (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1–3):1–41
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2006) The protein data bank, 1999–. In: International tables for crystallography Volume F: Crystallography of biological macromolecules. Springer, pp 675–684
Sali A (1994) Modeller. A program for protein structure modeling by satisfaction of spatial restraints. http://guitar.rockefeller.edu/modiller/modeller.html
Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61(4):704–721
Fakhar Z, Govender T, Maguire GE, Lamichhane G, Walker RC, Kruger HG, Honarparvar B (2017) Differential flap dynamics in l, d-transpeptidase2 from mycobacterium tuberculosis revealed by molecular dynamics. Mol BioSyst 13(6):1223–1234
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52(7):1757–1768
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1. Adv Drug Deliv Rev 46(1–3):3–26
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Singh UC, Kollman PA (1984) An approach to computing electrostatic charges for molecules. J Comput Chem 5(2):129–145
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36(22):3219–3228
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
BIOVIA DS (2017) BIOVIA Discovery Studio 2017 R2: A comprehensive predictive science application for the Life Sciences. San Diego, CA, USA http://accelrys.com/products/collaborative-science/biovia-discovery-studio
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) Epik: a software program for pK a prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21(12):681–691
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL (2015) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12(1):281–296
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267(3):727–748
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J Med Chem 49(21):6177–6196
Enyedy IJ, Egan WJ (2008) Can we use docking and scoring for hit-to-lead optimization? J Comput Aided Mol Des 22(3–4):161–168
Repasky MP, Shelley M, Friesner RA (2007) Flexible ligand docking with glide. Current protocols in bioinformatics:8.12. 11-18.12. 36
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21(6):1087–1092
Taylor RD, Jewsbury PJ, Essex JW (2002) A review of protein-small molecule docking methods. J Comput Aided Mol Des 16(3):151–166
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65(3):712–725
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174
Harvey M, De Fabritiis G (2009) An implementation of the smooth particle mesh Ewald method on GPU hardware. J Chem Theory Comput 5(9):2371–2377
Kräutler V, Van Gunsteren WF, Hünenberger PH (2001) A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem 22(5):501–508
John A, Sivashanmugam M, Umashankar V, Natarajan SK (2017) Virtual screening, molecular dynamics, and binding free energy calculations on human carbonic anhydrase IX catalytic domain for deciphering potential leads. J Biomol Struct Dyn 35(10):2155–2168
Tolufashe GF, Halder AK, Ibeji CU, Lawal MM, Ntombela T, Govender T, Maguire GE, Lamichhane G, Kruger HG, Honarparvar B (2018) Inhibition of mycobacterium tuberculosis L, D-transpeptidase 5 by carbapenems: MD and QM/MM mechanistic studies. ChemistrySelect 3(48):13603–13612
Silva JRA, Bishai WR, Govender T, Lamichhane G, Maguire GE, Kruger HG, Lameira J, Alves CN (2016) Targeting the cell wall of mycobacterium tuberculosis: a molecular modeling investigation of the interaction of imipenem and meropenem with L, D-transpeptidase 2. J Biomol Struct Dyn 34(2):304–317
Sun H, Li Y, Tian S, Xu L, Hou T (2014) Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys 16(31):16719–16729
Miller III BR, McGee Jr TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA. py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8(9):3314–3321
Islam MA, Pillay TS (2017) Identification of promising DNA GyrB inhibitors for tuberculosis using pharmacophore-based virtual screening, molecular docking and molecular dynamics studies. Chem Biol Drug Des 90(2):282–296
Martin YC (2005) A bioavailability score. J Med Chem 48(9):3164–3170
Bianchet MA, Pan YH, Basta LAB, Saavedra H, Lloyd EP, Kumar P, Mattoo R, Townsend CA, Lamichhane G (2017) Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase Ldt Mt2 by biapenem and tebipenem. BMC Biochem 18(1):8
Erdemli SB, Gupta R, Bishai WR, Lamichhane G, Amzel LM, Bianchet MA (2012) Targeting the cell wall of mycobacterium tuberculosis: structure and mechanism of L, D-transpeptidase 2. Structure 20(12):2103–2115
Hyndman RJ, Koehler AB (2006) Another look at measures of forecast accuracy. Int J Forecast 22(4):679–688
Lobanov MY, Bogatyreva N, Galzitskaya O (2008) Radius of gyration as an indicator of protein structure compactness. Mol Biol 42(4):623–628
Peterson K, Zimmt M, Linse S, Domingue R, Fayer M (1987) Quantitative determination of the radius of gyration of poly (methyl methacrylate) in the amorphous solid state by time-resolved fluorescence depolarization measurements of excitation transport. Macromolecules 20(1):168–175
Kassem S, Ahmed M, El-Sheikh S, Barakat KH (2015) Entropy in bimolecular simulations: a comprehensive review of atomic fluctuations-based methods. J Mol Graph Model 62:105–117. https://doi.org/10.1016/j.jmgm.2015.09.010
Chiba S, Harano Y, Roth R, Kinoshita M, Sakurai M (2012) Evaluation of protein-ligand binding free energy focused on its entropic components. J Comput Chem 33(5):550–560. https://doi.org/10.1002/jcc.22891
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng Des Sel 8(2):127–134
Funding
Our gratitude goes to Aspen Pharmacare, National Research Foundation (NRF), and the University of KwaZulu-Natal (UKZN) for the financial support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sabe, V.T., Tolufashe, G.F., Ibeji, C.U. et al. Identification of potent L,D-transpeptidase 5 inhibitors for Mycobacterium tuberculosis as potential anti-TB leads: virtual screening and molecular dynamics simulations. J Mol Model 25, 328 (2019). https://doi.org/10.1007/s00894-019-4196-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-019-4196-z