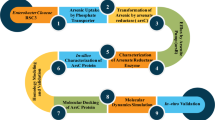
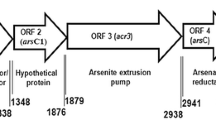
Abstract
Arsenic prevalence in the environment impelled many organisms to develop resistance over the course of evolution. Tolerance to arsenic, either as the pentavalent [As(V)] form or the trivalent form [As(III)], by bacteria has been well studied in prokaryotes, and the mechanism of action is well defined. However, in the rod-shaped arsenic tolerant Deinococcus indicus DR1, the key enzyme, arsenate reductase (ArsC) has not been well studied. ArsC of D. indicus belongs to the Grx-linked prokaryotic arsenate reductase family. While it shares homology with the well-studied ArsC of Escherichia coli having a catalytic cysteine (Cys 12) and arginine triad (Arg 60, 94, and 107), the active site of D.indicus ArsC contains four residues Glu 9, Asp 53, Arg 86, and Glu 100, and with complete absence of structurally equivalent residue for crucial Cys 12. Here, we report that the mechanism of action of ArsC of D. indicus is different as a result of convergent evolution and most likely able to detoxify As(V) using a mix of positively- and negatively-charged residues in its active site, unlike the residues of E. coli. This suggests toward the possibility of an alternative mechanism of As (V) degradation in bacteria.



Similar content being viewed by others
Data availability
The model used in this study has been deposited at ModelArchive (https://www.modelarchive.org/doi/10.5452/ma-abpir) and is available for public use. The trajectory snapshot from MD simulation has been uploaded as Supplementary material.
References
Nriagu JO, Pacyna JM (1988) Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 333(6169):134–139. https://doi.org/10.1038/333134a0
Nordstrom DK (2002) Public health. Worldwide occurrences of arsenic in ground water. Science 296(5576):2143–2145. https://doi.org/10.1126/science.1072375
Smith AH, Hopenhayn-Rich C, Bates MN, Goeden HM, Hertz-Picciotto I, Duggan HM, Wood R, Kosnett MJ, Smith MT (1992) Cancer risks from arsenic in drinking water. Environ Health Perspect 97:259–267
Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, Suk WA (2013) The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 121(3):295–302. https://doi.org/10.1289/ehp.1205875
Chen CJ, Hsueh YM, Lai MS, Shyu MP, Chen SY, Wu MM, Kuo TL, Tai TY (1995) Increased prevalence of hypertension and long-term arsenic exposure. Hypertension 25(1):53–60
Karagas MR, Tosteson TD, Blum J, Morris JS, Baron JA, Klaue B (1998) Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U. S. population. Environ Health Perspect 106(Suppl 4):1047–1050
Chu HA, Crawford-Brown DJ (2006) Inorganic arsenic in drinking water and bladder cancer: a meta-analysis for dose-response assessment. Int J Environ Res Public Health 3(4):316–322
Cullen WR, Reimer KJ (1989) Arsenic speciation in the environment. Chem Rev 89(4):713–764. https://doi.org/10.1021/cr00094a002
Liu SX, Athar M, Lippai I, Waldren C, Hei TK (2001) Induction of oxyradicals by arsenic: implication for mechanism of genotoxicity. Proc Natl Acad Sci U S A 98(4):1643–1648. https://doi.org/10.1073/pnas.031482998
Klaassen CD, Casarett LJ, Doull J (2013) Casarett and Doull's toxicology: the basic science of poisons, vol xiii, 8th edn. McGraw-Hill, New York
Kaltreider RC, Davis AM, Lariviere JP, Hamilton JW (2001) Arsenic alters the function of the glucocorticoid receptor as a transcription factor. Environ Health Perspect 109(3):245–251
Silver S, Phung LT (2005) Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol 71(2):599–608. https://doi.org/10.1128/AEM.71.2.599-608.2005
Rosen BP, Liu Z (2009) Transport pathways for arsenic and selenium: a minireview. Environ Int 35(3):512–515. https://doi.org/10.1016/j.envint.2008.07.023
Sanders OI, Rensing C, Kuroda M, Mitra B, Rosen BP (1997) Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. J Bacteriol 179(10):3365–3367
Rosen BP (1999) Families of arsenic transporters. Trends Microbiol 7(5):207–212
Carlin A, Shi W, Dey S, Rosen BP (1995) The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol 177(4):981–986
Diorio C, Cai J, Marmor J, Shinder R, DuBow MS (1995) An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol 177(8):2050–2056
Ji G, Silver S (1992) Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci U S A 89(20):9474–9478
Chauhan D, Srivastava PA, Yennamalli RM, Priyadarshini R (2017) Draft genome sequence of Deinococcus indicus DR1, a novel strain isolated from a freshwater wetland. Genome Announce 5(31). https://doi.org/10.1128/genomeA.00754-17
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34(Web Server issue):W369–W373. https://doi.org/10.1093/nar/gkl198
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32(Web Server issue):W526–W531. https://doi.org/10.1093/nar/gkh468
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. https://doi.org/10.1002/jcc.20291
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854. https://doi.org/10.1093/bioinformatics/btt055
Berendsen HJC, Vanderspoel D, Vandrunen R (1995) Gromacs - a message-passing parallel molecular-dynamics implementation. Comput Phys Commun 91(1–3):43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Jabeen A, Mohamedali A, Ranganathan S (2019) Protocol for protein structure modelling. In: Ranganathan S, Gribskov M, Nakai K, Schönbach C (eds) Encyclopedia of bioinformatics and computational biology. Academic, Oxford, pp 252–272. https://doi.org/10.1016/B978-0-12-809633-8.20477-9
Dwyer DS (2003) Molecular modeling and molecular dynamics simulations of membrane transporter proteins. In: Yan Q (ed) Membrane transporters: methods and protocols. Humana, Totowa, pp 335–350. https://doi.org/10.1385/1-59259-387-9:335
Kokabu Y, Ikeguchi M (2013) Molecular modeling and molecular dynamics simulations of recombinase Rad51. Biophys J 104(7):1556–1565. https://doi.org/10.1016/j.bpj.2013.02.014
Zimmermann MT, Urrutia R, Oliver GR, Blackburn PR, Cousin MA, Bozeck NJ, Klee EW (2017) Molecular modeling and molecular dynamic simulation of the effects of variants in the TGFBR2 kinase domain as a paradigm for interpretation of variants obtained by next generation sequencing. PLoS One 12(2):e0170822. https://doi.org/10.1371/journal.pone.0170822
Hermans J, Berendsen HJC, Vangunsteren WF, Postma JPM (1984) A consistent empirical potential for water-protein interactions. Biopolymers 23(8):1513–1518. https://doi.org/10.1002/bip.360230807
Postma JPM, Berendsen HJC, Straatsma TP (1984) Intramolecular vibrations from molecular-dynamics simulations of liquid water. J Phys-Paris 45(Nc-7):31–40. https://doi.org/10.1051/jphyscol:1984703
Buneman O (1983) Computer-simulation using particles - Hockney,Rw, Eastwood,Jw. SIAM Rev 25(3):425–426. https://doi.org/10.1137/1025102
Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR (1984) Molecular-dynamics with coupling to an external Bath. J Chem Phys 81(8):3684–3690. https://doi.org/10.1063/1.448118
Parrinello M, Rahman A (1981) Polymorphic transitions in single-crystals - a new molecular-dynamics method. J Appl Phys 52(12):7182–7190. https://doi.org/10.1063/1.328693
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105(28):6474–6487. https://doi.org/10.1021/jp003919d
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472. https://doi.org/10.1002/(Sici)1096-987x(199709)18:12<1463::Aid-Jcc4>3.0.Co;2-H
Wennberg CL, Murtola T, Hess B, Lindahl E (2013) Lennard-Jones lattice summation in bilayer simulations has critical effects on surface tension and lipid properties. J Chem Theory Comput 9(8):3527–3537. https://doi.org/10.1021/ct400140n
Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24(23):2780–2781. https://doi.org/10.1093/bioinformatics/btn507
Suresh K, Reddy GS, Sengupta S, Shivaji S (2004) Deinococcus indicus sp. nov., an arsenic-resistant bacterium from an aquifer in West Bengal, India. Int J Syst Evol Microbiol 54(Pt 2):457–461. https://doi.org/10.1099/ijs.0.02758-0
Mukhopadhyay R, Rosen BP (2002) Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect 110(Suppl 5):745–748
Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D (2013) High-resolution comparative modeling with RosettaCM. Structure 21(10):1735–1742. https://doi.org/10.1016/j.str.2013.08.005
DesJarlais RL, Sheridan RP, Seibel GL, Dixon JS, Kuntz ID, Venkataraghavan R (1988) Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J Med Chem 31(4):722–729
Laskowski RA, Luscombe NM, Swindells MB, Thornton JM (1996) Protein clefts in molecular recognition and function. Protein Sci 5(12):2438–2452. https://doi.org/10.1002/pro.5560051206
Acknowledgments
PAS, VA, and RMY acknowledge Jaypee University of Information Technology for providing research facilities to conduct this study. RP acknowledges Shiv Nadar University for providing research facilities and DC is supported by a doctoral research fellowship from Shiv Nadar University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Fig. S1
Ramachandran plot of model 1. (PNG 426 kb)
Supplementary Fig. S2
Root mean square deviation analysis of MD simulation. The least square fit of the entire protein from 0 ns was compared to snapshots of the entire protein at frequent intervals throughout the 100 ns simulation. The plot indicates equilibration of model 1 reached after 20 ns or so. (PNG 68 kb)
Supplementary Fig. S3
Root mean square fluctuation of residue of MD simulation. The fluctuation throughout the trajectory for each residue in model 1 is plotted, where the maximum fluctuation is observed in the C-terminal region of the protein. (PNG 194 kb)
Supplementary Fig. S4
Radius of gyration of model 1. The radius of gyration of the entire protein fluctuates between 1.4 nm to 1.52 nm throughout the trajectory indicating that the model is stable and does not denature during the simulation. (PNG 55 kb)
Supplementary Fig. S5
Molecular docking of As (III) with ArsC of D. indicus and residues that are within 5 Å radius. (PNG 2067 kb)
ESM 1
(DOCX 13 kb)
ESM 2
(DOCX 18 kb)
ESM 3
(PDB 14357 kb)
Rights and permissions
About this article
Cite this article
Chauhan, D., Srivastava, P.A., Agnihotri, V. et al. Structure and function prediction of arsenate reductase from Deinococcus indicus DR1. J Mol Model 25, 15 (2019). https://doi.org/10.1007/s00894-018-3885-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3885-3