Abstract
The synthesis and nuclease activity of a new bifunctional heterodinuclear platinum–copper complex are reported. The design of this ditopic coordination compound is based on the specific mode of action of each component, namely, cisplatin and Cu(3-Clip-Phen), where 3-Clip-Phen is 1-(1,10-phenanthrolin-3-yloxy)-3-(1,10-phenanthrolin-8-yloxy)propan-2-amine. Cisplatin is not only able to direct the Cu(3-Clip-Phen) part to the GG or AG site, but also acts as a kinetically inert DNA anchor. The nuclease activity of this complex has been investigated on supercoiled DNA. The dinuclear compound is not only more active than Cu(3-Clip-Phen), but is also capable of inducing direct double-strand breaks. The sequence selectivity of the mononuclear platinum complex has been investigated by primer extension experiments, which reveal that its interaction with DNA occurs at the same sites as for cisplatin. The Taq polymerase recognizes the resulting DNA damage as different from that for unmodified cisplatin. The sequence-selective cleavage has been investigated by high-resolution gel electrophoresis on a 36-bp DNA fragment. Sequence-selective cleavages are observed in the close proximity of the platinum sites for the strand exhibiting the preferential platinum binding sites. The platinum moiety also coordinates to the other DNA strand, most likely leading only to mono guanine or adenine adducts.
Similar content being viewed by others
Introduction
DNA is a target for numerous antitumor drugs [1, 2]. Reversible or irreversible modifications of the nucleic acids can lead to disruption of the transcription and/or replication, initiating ultimately the death of cancer cells. Cisplatin [3] and bleomycin are among these efficient anticancer drugs [4]. The mechanism of action is different for both drug substances; cisplatin primarily induces distortions upon binding to DNA, whereas bleomycin is able to generate DNA strand scissions.
Since the discovery of bleomycin [5], numerous metal complexes have been synthesized that are able to produce DNA cleavage. Typical examples are iron(II)–edta complexes, manganese(III)–porphyrin and Cu, Co, Ru and Rh complexes with phenanthroline [6–9]. For instance, Cu(phen)2 + in the presence of dihydrogen peroxide efficiently cleaves double-stranded DNA through the oxidative attack on deoxyribose units from the minor groove [10, 11]. The consequent DNA-cleavage products include 5′-monophosphate and 3′-monophosphate ester termini, free bases, 5-methylene furanone, and a small amount of 3′-phosphoglycolate [12–14]. Dihydrogen peroxide can be generated by Cu(phen)2 2+ in close proximity to the DNA strands, in the presence of a reductant and molecular oxygen [15]. The nuclease activity of Cu(phen)2 has been enhanced with the synthesis of 1-(1,10-phenanthrolin-3-yloxy)-3-(1,10-phenanthrolin-8-yloxy)propan-2-amine (3-Clip-Phen, 4) based on the covalent linkage of two phenanthroline units through their 3-position, leading to an increase of 60 times compared with the nuclease activity of Cu(phen)2 itself [15]. However, similarly to Cu(phen)2, the copper complexes of 3-Clip-Phen have no sequence selectivity, and cleave the DNA in a single-stranded fashion. The amine group of 3-Clip-Phen has been functionalized with different groups, such as a distamycin analog or various DNA intercalators [16, 17]. Thus, the resulting complexes show enhanced cleaving activities, and the complexes with the distamycin analog exhibit excellent targeting properties toward A·T boxes [18].
Recently, it has been shown that Cu(3-Clip-Phen) (3) attached to a cisplatin motif is able to perform direct double-strand cuts, thanks to the DNA-anchoring platinum moiety [19]. Platinum complexes have received considerable attention since the discovery of the antiproliferate activity of cisplatin in 1969 [20]. It is generally accepted that the distortion of DNA generated upon binding of cisplatin is largely responsible for its antitumor properties [3]. Subsequent drug activation via intracellular aquation reactions results in a variety of stable bifunctional DNA–platinum(II) adducts. 1,2-Intrastrand cross-links between two adjacent guanine bases d(GG) or between an adenine and a guanine residue d(AG) are primarily formed. The platinum center of complexes with a cis motif preferentially coordinates to the N7 position of both adenine and guanine in the major groove of DNA [21–24].
In the present paper, the preparation of a bifunctional complex [CuPt[N 1-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine]Cl4, 2] containing both a cis-bis(amine)Pt(II) moiety and a nuclease active 3-Clip-Phen group is reported (Fig. 1). The bridge connecting the platinum and copper moieties is very short and therefore rigid in contrast to that in the previous report, where the complexes had flexible linkers in order to have a major–minor groove interaction [19]. The platinum and the 3-Clip-Phen adjacent parts are now separated by a short linker. Consequently, in this case either the platinum species will not interact with its preferential site (the major groove) or the 3-Clip-Phen moiety will not interact with its preferential site (the minor groove). The platinum component plays two roles: (1) it acts as a DNA anchor, thus allowing the Cu(3-Clip-Phen) moiety to perform cleavages in the close proximity of the Pt–DNA adducts, and (2) it induces a sequence-selective binding of the heterodinuclear complex. Accordingly, the achievement of double-strand DNA breaks is potentially increased, because the single-strand cuts are in the close proximity of the platinum adduct and sequence-selective cleavage may be expected.
In the present study, the binding property of the platinum moiety and the cleavage selectivity and activity of this novel heterodinuclear complex 2 were investigated by agarose gel electrophoresis and high-resolution analysis with a 36-bp DNA fragment (Fig. 2). The results obtained are compared with those achieved with cisplatin and the Cu(3-Clip-Phen) complex.
Materials and methods
Preparation of Pt[N 1-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine]Cl2 and 2
All reagents and solvents were commercially available and used without further purification. Cu(3-Clip-Phen) (3) was prepared as previously described [15]. The 1H NMR chemical shifts of standard solvent molecules have been omitted in all redrawn spectra for clarity.
2-(2-(3-(1,10-Phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-ylamino)ethyl)isoindoline-1,3-dione
3-Clip-Phen (4) (100 mg, 0.22 mmol) was dissolved in 5 mL of dimethylformamide (DMF). One equivalent of N-(2-bromoethyl)phthalimide (5) (56.7 mg, 0.22 mmol) and 1 equiv of N-ethyldiisopropylamine (38 μL, 0.22 mmol) were added to 4. The reaction mixture was heated to 100 °C for 2 days. After the mixture had cooled to room temperature, 10 mL of dichloromethane (DCM) was added, and the organic phase was washed three times with 10 mL of distilled water. After being dried over Na2SO4 and evaporation under reduced pressure, the resulting crude product was purified by column chromatography (SiO2, DCM, MeOH, NH4OH, 95:5:0.5), to yield 2-(2-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-ylamino)ethyl)isoindoline-1,3-dione (6) as an off-white powder (yield 34%). 1H NMR (DCCl3, 300 MHz) δ 9.05 (dd, 2H, J = 4.32, 1.67 Hz), 8.81 (d, 2H, J = 2.83 Hz), 8.10 (dd, 2H, J = 8.07, 1.62 Hz), 7.72 (dd, 2H, J = 5.43, 3.05 Hz), 7.67 (d, 2H, J = 8.88 Hz), 7.62 (d, 2H J = 8.86 Hz), 7.54 (dd, 2H, J = 6.09, 4.74 Hz), 7.48 (m, 4H), 4.27 (d, 4H, J = 5.18 Hz), 3.84 (t, 2H, J = 6.06 Hz), 3.54 (m, 1H), 3.13 (t, 2H, J = 6.12 Hz) ppm. 13C NMR (DCCl3, 75 MHz) δ 168.4, 153.7, 150.1, 146.0, 142.4, 140.3, 135.73, 133.8, 132.0, 129.3, 127.1, 125.9, 123.0, 122.0, 114.9, 67.3, 55.6, 45.4, 37.8 ppm. Low-resolution mass spectrometry (MS) (electrospray ionization, ESI, more than 0) m/z 621.00 [(M + H)+; calcd for C37H29N6O4 +: 621.66], 642.94 [(M + Na)+; calcd for C37H28N6O4Na+: 643.65] Anal. calcd for C37H28N6O4·1.6 H2O: C, 68.42; H, 4.84; N, 12.94. Found: C, 68.34; H, 4.86; N, 13.21.
N 1-(3-(1,10-Phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine
Two equivalents of hydrazine (15.6 μL, 0.32 mmol) were added to a solution of 6 (100 mg, 0.16 mmol) in 5 mL of pure ethanol. The mixture was refluxed overnight. The ethanol was evaporated under reduced pressure, and the crude product was purified by column chromatography (SiO2, DCM, MeOH, NH4OH, 90:10:1) to give N 1-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine (7) as a light-brown powder (yield 73%). 1H NMR (MeOD-d 3, 300 MHz) δ 9.00 (dd, 2H, J = 4.41, 1.63 Hz), 8.84 (d, 2H, J = 2.84 Hz), 8.34 (dd, 2H, J = 8.10, 1.67 Hz), 7.90 (d, 2H, J = 2.86 Hz), 7.83 (m, 4H), 7.65 (dd, 2H, J = 8.10, 3.26 Hz), 4.51 (d, 4H, J = 4.47 Hz), 3.58 (m, 1H), 3.07 (t, 2H, J = 5.64 Hz), 2.97 (t, 2H, J = 5.23) ppm. 13C NMR (MeOD-d 3, 75 MHz) δ 155.6, 150.6, 146.3, 143.0, 140.5, 137.6, 131.0, 128.7, 128.2, 127.4, 123.5, 116.7, 69.1, 57.5, 41.9 ppm. Low-resolution MS (ESI > 0) m/z 490.98 [(M + H)+; calcd for C29H27N6O2 +: 491.56] Anal. calcd for C29H26N6O2·1.9 CH2Cl2: C, 56.93; H, 4.61; N, 12.89. Found: C, 56.79; H, 5.13; N, 13.23.
Pt[N 1-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine]Cl2
To a solution of 7 (70.4 mg, 0.14 mmol) in 6.5 mL of DMF was added a solution of 0.8 equiv of K2PtCl4 (47.69 mg, 0.11 mmol) in 3.25 mL of de-ionized H2O. The reaction mixture was stirred for 6 h at room temperature. The off-white precipitate was filtered off and washed with 30 mL of deionized water, 30 mL of MeOH and 20 mL of diethyl ether to give Pt[N 1-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine]Cl2 (1) as an off-white powder (yield 42%). 1H NMR (DMSO-d 6, where DMSO is dimethyl sulfoxide, 300 MHz) δ 9.04 (d, 2H, J = 2.77 Hz), 8.88 (d, 2H, J = 2.45 Hz), 8.45 (d, 2H, J = 8.00 Hz), 8.05 (br, 2H), 7.95 (m, 4H), 7.70 (dd, 2H, J = 7.99, 4.35 Hz), 4.49 (br, 4H), 4.07 (br, 5H) ppm. 195Pt NMR (DMSO-d6) δ −2,317 (complex), −3,292 (complex with one DMSO coordinated) ppm. IR (neat): ν = 3,049 (br); 1,591 (s); 1,506 (s); 1,424 (s); 1,236 (s); 1,198 (s); 1,102 (s); 1,018 (s); 876 (s), 834 (s), 728 (s), 332 (s), 325 (s) cm−1. Anal. calcd for C29H26Cl2N6O2Pt·DMF: C, 46.33; H, 4.01; N, 11.82 Found: C, 47.54; H, 4.43; N; 11.70; the deviation in calculated percentage C is ascribed to traces of solvent other than DMF.
CuPt[N1-(3-(1,10-phenanthrolin-3-yloxy)-1-(1,10-phenanthrolin-8-yloxy)propan-2-yl)ethane-1,2-diamine]Cl4
CuCl2 (17.0 mg, 0.1 mmol) was added as a solid to a suspension of 1 (75.7 mg, 0.1 mmol) in DMF (25 mL). The reaction was stirred overnight at 50 °C. DMF was partially evaporated under reduced pressure, and the crude compound was precipitated in 100 mL of diethyl ether. The solid material was filtered off and washed with 3 × 20 mL of diethyl ether, and dried overnight at 50 °C under reduced pressure to give 2 as a green powder (yield 93%). X-band electron paramagnetic resonance (solid state): g = 2.1191. UV–vis (H2O) λ max 282 (40,100), 321 (12,500), 333 (10,100), 347 (3,280) nm. High-resolution MS (m/z): [2M–2Cl] calcd for (C58H52Cl6N12O4Pt2Cu2)2+ 853.00852, 853.5096, 854.0097, 854.5087, 855.0088, 855.5072, 856.0073, 856.5083, 857.0100, 857.5067; found 853.0117, 853.5117, 854.0114, 854.5110, 855.0107, 855.5105, 856.0103, 856.5101, 857.0098, 857.5097.
Cytotoxicity
The cytotoxicity of complexes 1 and 2 has been determined for breast (MCF7), two glioblastoma (Hs683 and U373), two colorectal (HCT-15 and LoVo) and lung (A549) cancer cell lines (see supplementary material for experimental details and results).
Solutions of complexes for experiments with DNA
One millimolar DMSO solutions of the complexes investigated were prepared, and were subsequently diluted with Milli-Q water.
Nuclease activity on supercoiled DNA
One millimolar DMSO solutions of the complexes investigated were diluted to, respectively, 200, 400 and 1,000 nM with Milli-Q water. Five microliters of the complex solution was added to 10 μL of supercoiled ΦX174 DNA (Invitrogen, 7 nM, 40 μM base pairs) in 6 mM NaCl, 20 mM sodium phosphate buffer (pH 7.2), and incubated for 20 h at 37 °C. To initiate the cleavage, 5 μL of a 20 mM mercaptopropionic acid solution in water was added, and the resulting reaction mixture was incubated at 37 °C for 1 h. The reaction was quenched at 4 °C, followed by the addition of 4 μL of loading buffer (glycerol with bromophenol blue) prior to the loading on a 0.8% agarose gel containing 1 μg mL−1 ethidium bromide. The gels were run at a constant voltage of 70 V for 90 min in 2-amino-2-(hydroxymethyl)propane-1,3-diol (Tris) borate edta (TBE) buffer containing 1 μg mL−1 ethidium bromide. The gels were visualized under a UV transilluminator, and the bands were quantified using a Bio-Rad Gel Doc 1000 apparatus interfaced with a computer.
Time-course experiments of DNA cleavage
Fifty microliters of the complex solution was added to 100 μL of supercoiled ΦX174 DNA (Invitrogen, 7 nM, 40 μM base pairs) in 6 mM NaCl, 20 mM sodium phosphate buffer (pH 7.2), and the resulting reaction mixture was incubated for 20 h at 37 °C. To initiate the cleavage, 50 μL of 20 mM mercaptopropionic acid was added, and a sample was taken out every 10 min. Four microliters of loading buffer (glycerol with bromophenol blue) was added, and the sample was directly frozen in liquid nitrogen. When all samples had been collected, they were loaded on a 0.8% agarose gel containing 1 μg mL−1 ethidium bromide.
Analyses with 5′-32P-end-labeled DNA
Oligonucleotides (ODNs) I and II and the primer (Fig. 2) were purchased from Eurogentec, and purified on a 15% polyacrylamide (acrylamide to bisacrylamide ratio 19:1) gel. The denaturating gel contained 7 M urea. The migrations was performed in the presence of TBE buffer at 2,500 V for 3 h. The concentrations of single-stranded ODNs were determined by UV titration at 260 nm [25]. The ODNs were end-labeled with 32P using standard procedures with T4 polynucleotide kinase (New England BioLabs) and [γ-32P]ATP for the 5′-end, before being purified on a MicroSpin G25 column (Pharmacia) [26].
Comparison of the platinum–ODN adducts formed with the different complexes
5′-end-labeled ODN I (2 μM) was annealed to 1 equiv of its complementary strand ODN II in 1,100 μL of Tris–HCl (20 mM, pH 7.2) by heating to 90 °C for 5 min, followed by slow cooling to room temperature. Then, 60 μL of this solution was incubated with 60 μL of the complex solution (6 or 20 μM) for 20 h at 37 °C, followed by precipitation with 100 μL of sodium acetate buffer (3 M, pH 5.2) and 1,300 μL of cold ethanol. Pellets were rinsed with ethanol and lyophilized. Samples were dissolved in formamide containing bromophenol blue and xylene cyanol and heated for 3 min at 90 °C before being loaded on the gel. Platinum–DNA adducts were analyzed by denaturing 20% polyacrylamide gel electrophoresis, then by phosphorimagery.
Comparison of the sequence-selective binding by primer extension experiments with Taq polymerase
ODN I (2 μM) was annealed to ODN II (2 μM) in 1,100 μL of Tris–HCl (20 mM, pH 7.2) by heating to 90 °C for 5 min, followed by slow cooling to room temperature. Sixty microliters of this solution was then incubated with 60 μL of the complex solution (6 or 20 μM) for 20 h at 37 °C, followed by precipitation with 100 μL of sodium acetate buffer (3 M, pH 5.2) and 1,300 μL of cold ethanol. Pellets were rinsed with ethanol and lyophilized. For primer extension, an aliquot of the redissolved samples (0.25 μM) was annealed with 5′-end-labeled primer (0.25 μM) and 1 equiv of ODN I (0.25 μM) in the enzyme buffer (the buffer contains a small amount of reductant, but not enough to induce DNA cleavage by complex 2) before the addition of 250 μM dGTP, dCTP, dATP and dTTP and 2.5 U of Taq polymerase (final concentrations are given, the total volume was 10 μL). One equivalent of unmodified ODN I was added in order to displace the ODN II from the duplex and replace it with the labeled primer. The samples were reacted at 37 °C for 30 min, and 1 μL of edta (0.2 M) was subsequently added. Samples were dissolved in formamide containing bromophenol blue and xylene cyanol and heated for 3 min at 90 °C, before being loaded on the gel. Five microliters of sample was then analyzed by denaturing 20% polyacrylamide gel electrophoresis and phosphorimagery. The Maxam and Gilbert [27] sequencing scale, including a final scale of T4 polynucleotide kinase digestion to remove 3′-end phosphates, was used to analyze the DNA fragments. The sequencing experiments show, in addition to a strong, easily assigned G signal, also a weak A signal.
Comparison of the cleavage patterns of ODN I–ODN II induced by the copper complexes
The 5′-end-labeled 36mer target (2 μM) was annealed to 1 equiv of its complementary strand in 1,100 μL of Tris–HCl (20 mM, pH 7.2) by heating to 90 °C for 5 min, followed by slow cooling to room temperature. To 60 μL of this solution was added 60 μL of complex 1, 2 or 3 solutions (20 μM). Some of the samples involving complex 2 were incubated for 19 h, followed by the addition of 1 equiv of CuCl2 per complex and subsequent incubation (1 h). The other samples were incubated for 20 h at 37 °C. Next, all samples were precipitated with 100 μL of sodium acetate buffer (3 M, pH 5.2) and 1,300 μL of cold ethanol. Pellets were rinsed with ethanol and lyophilized, then dissolved to 1.33 μM in Tris–HCl buffer (13.3 mM, pH 7.4). For the cleavage experiments, to 15 μL of this solution was added 5 μL of a 0.8 mM ascorbate solution (5 μL of water was added to the controls). The samples were incubated at 37 °C for 1 h, followed by precipitation in 20 μL of sodium acetate buffer (3 M, pH 5.2) containing 1 μg of salmon testes DNA and 180 μL of cold ethanol. Pellets were rinsed with ethanol and lyophilized. In order to study the DNA cleavage mechanism, additional treatments were performed on some samples: (1) heating at 90 °C in 50 μL of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–NaOH buffer (0.1 M, pH 8.0) for 30 min, followed by ethanol precipitation; (2) heating at 90 °C in 50 μL of piperidine (0.2 M in water) for 30 min, followed by lyophilization. Samples were dissolved in formamide containing bromophenol blue and xylene cyanol and heated for 3 min at 90 °C, before being loaded on the gel. Samples were analyzed by denaturing 20% polyacrylamide gel electrophoresis, then phosphorimagery. The Maxam and Gilbert [27] sequencing scale was used to analyze DNA fragments. The sequencing experiments show, in addition to a strong, easily assigned G signal, also a weak A signal.
Results and discussion
Design and synthesis of a heterodinuclear platinum–copper complex
A ditopic ligand 7 was designed to favor the simultaneous coordination of a platinum and a copper entity; hence, the resulting bifunctional complex would combine the ability to form a kinetically inert coordination bond with DNA, thanks to its cis-Pt moiety, with the cleavage properties of Cu(3-Clip-Phen). Two coordination compounds, namely, the platinum complex 1 and the heterodinuclear platinum–copper complex 2 (Fig. 1) were prepared with the ligand 7. The binding ability of the platinum unit and the cleavage selectivity of the Cu(3-Clip-Phen) moiety were investigated. The results were compared with those for the known Cu(3-Clip-Phen) complex (3). The short separation between the Pt and the Cu centers in complex 2 most likely affects the preference for platinum to bind in the major groove, and in the minor groove for copper. Accordingly, the coordination of the platinum moiety in the major groove will force the Cu(3-Clip-Phen) component to bind in the major groove, and vice versa.
The general synthetic pathway to prepare complexes 1 and 2 is depicted in Scheme 1. The selective and complete platination of the ethylenediamine unit using 1 equiv of K2PtCl4 was monitored by 195Pt NMR and 1H NMR. The in situ reaction of the resulting platinum derivative 1 with 1 equiv of CuCl2 produces the heterobimetallic complex 2.
Cleavage of supercoiled DNA
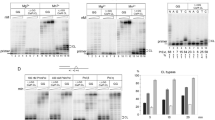
The relaxation of supercoiled circular ΦX174 DNA (form I) into its relaxed (form II) and linear (form III) conformations was monitored to compare the aerobic cleavage abilities of complexes 2 and 3, in the presence of a reducing agent (Fig. 3). First, the complexes were incubated for 20 h, to allow the formation of platinum–DNA adducts. The nuclease activity was subsequently initiated by the addition of 5 mM mercaptopropionic acid. Complex 2 exhibits a markedly higher nuclease activity than Cu(3-Clip-Phen) (3) (compare lanes 5 and 9 in Fig. 3). Indeed, at complex concentrations of 100 nM, most supercoiled DNA has reacted to form circular and linear DNA in the case of 2, while Cu(3-Clip-Phen) only generates a small amount of form II (Fig. 3, lanes 4 and 8). Moreover, almost all the DNA has reacted to form smaller DNA fragments (migrating as a smear) at a complex concentration of 250 nM for 2 (Fig. 3, lane 5), whereas no smear is observed for Cu(3-Clip-Phen) (Fig. 3, lane 9). Interestingly, at a concentration of 100 nM for complex 2, form III is detected before the total disappearance of form I (Fig. 3, lane 4) [19]. This result indicates that the heterodinuclear platinum–copper complex is able to perform direct double-strand cuts since form I is still detected in the reaction. Such double-strand breaks are highly cytotoxic, since the cells have difficulties to repair such damage [28, 29]. Cu(3-Clip-Phen) is only able to perform repetitive single-strand cuts [15]. Interestingly, the cleavage activity of complex 2 is also more efficient compared with that of the platinum–copper complexes previously reported by our group [19].
Comparative experiments of the oxidative cleavage of ΦX174 plasmid DNA performed by 2 and 3. Lane 1 control DNA. Lane 2 250 nM solution of 2. Lane 3 50 nM 2, in the presence of 5 mM mercaptopropionic acid (MPA). Lane 4 100 nM 2, in the presence of 5 mM MPA. Lane 5 250 nM 2, in the presence of 5 mM MPA. Lane 6 250 nM 3. Lane 7 50 nM 3, in the presence of 5 mM MPA. Lane 8 100 nM 3, in the presence of 5 mM MPA. Lane 9 250 nM 3, in the presence of 5 mM MPA
Time-course studies of DNA cleavage by complexes 2 and 3 were carried out to further investigate the direct double-strand cleavage event (Fig. 4). Complex 3 at a concentration of 250 nM generates a maximum of 80% of circular DNA (form II) after a reaction time of about 1 h [19]. Around 20% of linear DNA (form III) is produced via the action of 3 after 70 min. Remarkably, the formation of form III is only observed after a reaction time of 30 min when already 60% of form II has been produced. For a 150 nM solution of complex 2, linear DNA (form III) is generated from the initial stages of the cleavage reaction, with supercoiled DNA (form I) still being present (Fig. 4). Also, only a maximum of 60% of form II is generated. These features reflect the ability of 2 to perform direct double-strand breaks, most likely as a result of the binding of the platinum moiety to DNA, allowing the copper part to achieve more than one oxidative cleavage in the close proximity of the platinum coordination site.
Analysis of the platinum adducts on ODN I of the ODN I–ODN II duplex target
High-resolution analyses with a 36-bp DNA duplex (ODN I–ODN II) were performed to investigate the coordination of the platinum component of the bifunctional complexes to DNA, (Fig. 5a). The sequence of this duplex was chosen to include GG and AG sites (which are the two major binding sites of cis-Pt(II) complexes) on one strand (ODN I). The results obtained after incubation of the complexes with the duplex labeled on the 5′-end of ODN I for 24 h were analyzed by polyacrylamide gel electrophoresis under denaturing conditions and are shown in Fig. 5a. Denaturing conditions allowed the analysis of the Pt–DNA adducts on ODN I of the duplex.
a Polyacrylamide gel electrophoresis (PAGE) analysis of the Pt–ODN I adducts of the ODN I–ODN II duplex target (1 μM). ODN I was 5′-end-labeled with 32P-phosphate. The complexes were incubated with the DNA for 24 h before analyses. Lane 1 ODN I. Lane 2 3 μM cisplatin. Lane 3 10 μM cisplatin. Lane 4 3 μM 1. Lane 5 10 μM 1. Lane 6 3 μM 2. Lane 7 10 μM 2. b Phosphorimager data of a DNA sequencing gel comparing the sequence specificity of cisplatin, 1 and 2. All the samples were extended using Taq polymerase, starting from the 5′-end-labeled primer. Lane 1 blank experiment; Lane 2 3 μM cisplatin; Lane 3 10 μM 1. Lane 4 10 μM 2. It is noteworthy that the GTA and GGAC sites give the sequence of the opposite strand that induced the stopping of the primer extension (Figs. S1, S2). c Nucleobase sequence of ODN I, and indication of the damage sites induced by cisplatin, 1 and 2. Large arrows and small arrows represent, respectively, major and minor stop sites
Molecules able to irreversibly bind to DNA will retard the rate of migration of the modified ODN, compared with that of the free ODN; therefore, ODN–Pt adducts appear as retarded bands on gels. The incubation with 3 equiv of cisplatin clearly reveals an impeded mobility of the ensuing cisplatin–ODN I adduct (Fig. 5a, lane 2). Only traces of free ODN I were detected; 89% of the ODN I had been modified. The incubation of the DNA duplex with 10 equiv of cisplatin results in a total conversion of ODN I (Fig. 5a, lane 3). Complex 1 shows the formation of Pt–ODN I adducts (Fig. 5a, lanes 4 and 5). Two distinct bands are clearly observed when 10 equiv of complex 1 is incubated with the DNA target, indicating the formation of Pt–ODN I adducts. The quantification of free ODN I reveals that as much as 84% of this DNA fragment has reacted. The use of complex 2 leads to comparable results, with the conversion of 88% of the initial ODN I (Fig. 5a, lane 7). However, the reaction between ODN I and complex 2 produces a smear (i.e., a range of products) on the gel. Our previously reported compounds showed reduced formation of Pt–ODN adducts under similar experimental conditions compared with complexes 1 and 2 [19].
Sequence selective binding of complexes 1 and 2 compared with cisplatin
Primer extension experiments were performed to investigate the sequence-selective binding of the platinum units to the ODN I fragment of the ODN I–ODN II DNA duplex (Figs. 5b, c, S1, S2). The platinum complexes that did not react with DNA were removed prior to the start of the primer extension experiments. Nevertheless, it is possible to have more than one platinum moiety coordinating the ODN strand, since the concentrations of cisplatin, complex 1 and complex 2 were, respectively 3 and 10 times higher than the concentration of the ODN I–ODN II DNA duplex. Taq polymerase has proven to be a valuable tool for the determination of the sequence selectivity of various platinum complexes [23, 30–39]. Cisplatin inhibits the enzymatic polymerization at the anticipated GG and AG sites (Fig. 5b, c), but the majority of cisplatin is detected at the GG site. It should be noted that once the ODN I strand contains two Pt adducts on both the GG and AG site, the enzyme stops only at the GG site. Therefore, it is possible that the amount of modified AG sites is underestimated. Nevertheless, not all of the ODN I has been modified by the complexes (Fig. 5a). It is therefore reasonable to say that the majority of the ODN I contains only one Pt adduct and that the AG site is indeed the minor site of interaction. The stop sites observed for complex 1 are also located for a major part at the GG base pairs and for a minor part at the AG base pairs. Complex 2 shows only stops at the GG binding site. These results indicate that the platinum moiety interacts with its preferential binding site. The difference between cisplatin and complexes 1 and 2 is the precise point at which the peak intensity occurs at the damaged site. The stops of the Taq polymerase are mainly located at the A base before the GG site for cisplatin, complex 1 and complex 2. However, the enzyme stops induced at the GG site are more equally distributed among the GAC site in the cases of complexes 1 and 2 (Fig. 5b, c; the size of the arrows is an indication of the damage intensity). The binding of complexes 1 and 2 apparently produces bulky adducts, thus allowing a partial stop at the second nucleotide before the classic position, at the GG adduct, since an increase of the reaction time with the enzyme (from 30 to 120 min, Fig. S3; complex 1) induces a bypass of the stop at the C base (associated with a decrease of the peak intensity when compared with that for G). The difference between cisplatin and complex 1 is more pronounced at the AG site. Although it is the minor binding site, the damage induced by cisplatin almost exclusively takes place at the G base, at the 5′-end of ODN I. The Taq polymerase stops for complex 1 are mainly observed at the A base.
Cleavage of the ODN I strand of the ODN I–ODN II duplex
The cleavage of the ODN I–ODN II duplex with complexes 1–3 was investigated by polyacrylamide gel electrophoresis, the target being labeled on the 5′-end of ODN I (Fig. 6). To the best of our knowledge, studies of such bifunctional complexes have been reported only twice [40, 41].
a PAGE analysis of cleavage of ODN I of the ODN I–ODN II duplex (1 μM) by compounds 1–3 (10 μM when unspecified). The cleavage reactions were initiated with ascorbate (200 μM) in aerobic conditions or by heating for 30 min at 90 °C in aqueous 0.2 M piperidine. The Maxam–Gilbert sequencing reactions A + G (lane 1) and G (lane 2) were performed to determine the cleavage sites. On the top of the gel are indicated the conditions used during the experiments (details are given in “Materials and methods”). Lanes 3–6 are for controls without complexes. Lanes 17–22 are for experiments that were performed in the presence of 1 μM 3. b High-contrast picture of lanes 8, 10, 11, 13, 15 and 16 in a allowing easier observation of the the cleavage pattern. ΔpH 8 was a heating step of 30 min at 90 °C in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–NaOH buffer (0.1 M, pH 8.0). Unlinked complexes 1 and 2 were removed in a precipitation step with ethanol before the induction of cleavage
Complexes 1 and 2 were preincubated for 20 h to allow the coordination of the platinum moiety to the DNA target. For complex 1, the preincubation was subsequently followed by the coordination of 1 equiv of copper, since the copper-free complex does not show any nuclease activity. The noncoordinated complexes were removed by precipitation before the cleavage was initiated by the addition of ascorbic acid (0.2 mM) under aerobic conditions. Additional treatments (HEPES pH 8.0 and piperidine) were carried out.
The noncovalent interaction of Cu(3-Clip-Phen) (3) with DNA permits its abstraction from the ODNs by a simple precipitation step. A tenfold excess of 3 (compared with the ODNs) does not generate cleavage products after the precipitation step (Fig. 6, lane 24), while 1 equiv of 3 without precipitation exhibits significant cleavage (Fig. 6, lane 20). The platinum-containing complexes 1 and 2 form kinetically inert bonds with DNA, and therefore cannot be removed from the ODN during the precipitation step. In contrast, the unreacted complexes are eliminated.
As expected, the cleavage pattern of 3 is nonspecific, [18] as about 20% of the target is oxidized at all nucleotides (Fig. 6, lane 20). The cleavage achieved by complex 2 in the presence of ascorbic acid results in the full and partial disappearance of, respectively, the Pt–ODN I adducts and the ODN I band (Fig. 6, lane 10). The total cleavage amounts to nearly 80%, and its pattern is different from the features resulting from the action of 3, but no clear sequence selectivity is observed. Accordingly, a smear of products is noted in contrast to 3, suggesting that the mechanism of the cleavage is different. This observation may be explained by either of the following possibilities: (1) some cuts are due to platinum adducts of complex 2 positioned on the other strand of the duplex (ODN II); (2) the extensive cleavage observed most likely indicates that the ODN I fragment experiences more than one cut; therefore, complex 2 should have been released after repetitive cuts from ODN I and should be able to cut the free ODN.
To verify this hypothesis, the experimental conditions were adjusted to introduce fewer platinum adducts onto the target (20% against the 80% of modification of ODN I shown in Fig. 6), and less ascorbate (100 μM) was used to limit the recleavage events. However, the resulting cleavage pattern appears to be similar to that observed with the former conditions (Fig. 8).
One equivalent of CuCl2 was added to complex 1, after the formation of Pt–DNA adducts, to analyze the cleavage selectivity of the resulting compound. A comparatively less efficient cleavage was achieved using complex 1 with extra added CuCl2, when compared with complex 2 (compare lanes 10 and 15 in Fig. 6), possibly as a result of the partial coordination of the copper ions to 1. Similarly to 2, the cleavage pattern obtained with 1 is found to be significantly different from that of 3 (Fig. 6, lane 20). Interestingly, the free ODN I is not affected during the cleavage process, as only the ODN fragments containing platinum adducts are cleaved. Furthermore, a sequence-selective cleavage (associated with the formation of fragments of ODN I including probably the 3′-phosphate end, since they comigrated as Maxam and Gilbert sequencing fragments) is observed in the close vicinity of the GG and AG sites (indicated as black bars in Fig. 6, left). The intensity of the bands is much stronger at the GG site, reflecting the results of the primer extension experiments previously obtained. The four base pairs neighboring the GG site in the 5′-direction are also affected by the cleavage of complex 1. In the 3′-direction, no clear bands are observed, but a smear is observed, which can be explained as follows: the cleavage products remain coordinated to the complex and have therefore a totally different mobility, compared with the Maxam–Gilbert sequencing fragments. The apparent smear observed in the 3′-direction supports this assumption, since such behavior is observed for Pt–ODN I adducts including the copper complex (Fig. 5a, lane 6), and can therefore be expected for cleavage fragments that coordinate to the complex.
A heating step in HEPES buffer (90 °C; pH 8.0) is often used to cleave the metastable products resulting from the oxidation of deoxyribose. This treatment of the cleavage products obtained with complexes 1–3 did not show a strong increase of the DNA cuts (Fig. 6, respective comparisons between lanes 15, 16 and 10 and 11, 20 and 22). Therefore, products of strand cleavage are essentially observed during this analysis. However, the smears observed in lanes 10 and 15 are partially converted into bands, indicating that some cleavage products, arising from the oxidation of deoxyribose, are sensitive to alkaline conditions.
More drastic alkaline heating (as with piperidine) allows one to detect the oxidations of the nucleobases that do not induce direct DNA cleavage [6]. Surprisingly when this treatment is applied to complexes 1 (with addition of 1 equiv of copper) and 2, clear cleavages are observed, although the systems had not been incubated with ascorbate (Fig. 6, lanes 13 and 8, respectively). This phenomenon is dependent of the presence of copper ion, since it is not observed for Pt–ODN I adducts with complex 1 without the addition of CuCl2 (results not shown). Therefore, these alkaline conditions are sufficient to induce a redox activity of the copper complex part of the hybrid molecule [this phenomenon has not been observed with Cu(3-Clip-Phen), which can be removed during a precipitation step preceding the heating in the presence of piperidine]. Interestingly, only the Pt–ODN I adducts are cleaved; none of the free ODN I is degraded and selective cleavages are observed around the position of adducts detected during primer extension experiments. Unfortunately, this activity of the copper complexes covalently linked to the target does not allow us to analyze whether or not the dual Pt–Cu complexes perform the oxidation of nucleobases.
Cleavage of the ODN II strand of the ODN I–ODN II duplex
The same experiments were performed with the ODNI–ODNII duplex labeled on the 5′-end of ODN II (Fig. 7). Similar amounts of Pt–ODN II adducts are formed with complexes 1 (Fig. 7, lane 8) and 2 (Fig. 7, lane 3). Most likely, only parts of the platinum components of complexes 1 and 2 are bound on ODN II at the preferential AG site, and the cleavages do not seem to be restricted to one selective site. Other adducts are probably also present. The occurrence of platinum–guanine monoadducts on ODN II (which is particularly rich in guanine bases) may explain the results observed. Further investigations are required to confirm these proposals.
PAGE analysis of cleavage of ODN II of the ODN I–ODN II duplex (1 μM) by compounds 1–3. The cleavage reactions were initiated with ascorbate (200 μM) in aerobic conditions or by heating for 30 min at 90 °C in aqueous 0.2 M piperidine. The Maxam–Gilbert sequencing reactions A + G (lane 1) and G (lane 2) were performed to determine the cleavage sites. On top of the gel are indicated the conditions used during the experiments (details are given in “Materials and methods”). ΔpH 8 was a heating step of 30 min at 90 °C in HEPES–NaOH buffer (0.1 M, pH 8.0). Unlinked complexes 1 and 2 were removed in a precipitation step with ethanol before the induction of cleavage
Both complex 1 with added copper and complex 2 show extensive cleavage activities in the presence of ascorbic acid in air (Fig. 7, lanes 11 and 6, respectively). In the experiments with complex 2, the Pt–ODN II adducts have fully reacted, while the free ODN II is only partly altered. The cleavage fragments are poorly resolved on the gel (a smear is essentially observed), and treatment with HEPES or piperidine fails to improve their analysis. These cleavage fragments are composed of modified DNA. It can be reasonably proposed that the fragments include Pt–ODN II adducts and that these adducts produce a smear during the migration in polyacrylamide gel electrophoresis. All these results indicate that the platinum moieties of complexes 1 and 2 bind, as expected, to the preferential platinum site on ODN II, but also and mainly to single A and G nucleobases.
Comparison between the cleavage of duplex and single-stranded DNA
A comparison between the cleavage patterns resulting from the action of complex 2 on single-stranded ODN I and on the duplex ODN I–ODN II was made to appraise the influence of the formation of Pt–ODN II adducts on the resulting cleavage (Fig. 8). Pt–ODN I adducts are formed when the single strand is used, and these adducts are essentially positioned at the GG site of ODN I (the results of the primer extension are not shown). For this study, only 20% of the platinum adducts on ODN I of the duplex and 40% of the platinum adducts on the single strand were created in order to have a maximum of one adduct per DNA target, and thus to simplify the analysis. The cleavage was induced by the addition of a small quantity of ascorbate (100 μM) to disfavor recleavage events.
Phosphorimager scanning of the PAGE-cleavage patterns of ODN I by complex 2. The cleavage was performed on either the ODN I–ODN II duplex (5′-end-labeled on ODN I) or on single-stranded ODN I. Cleavage was induced by the addition of ascorbate (100 μM) after the removal of the unlinked complex during a precipitation step with ethanol. The clear band area on the 3′-side of the GG site of the duplex is probably due to a covalent adduct of complex 2 on ODN II (the complementary strand of the duplex). 3′-Phosphoglycolate cleavage fragments are labeled with an asterisk. The other clear bands were attributed to fragments of ODN I including 3′-phosphate ends
The cleavage of the ODN I fragment of the duplex results in a cleavage pattern exhibiting various moderate peaks in the 3′-region of the GG base pair of ODN I. The cleavage of single-stranded ODN I by complex 2 gives rise to a smear in the same region. The smears observed characterize cleaved ODN I products that are still coordinated to complex 2. Accordingly, the weak peaks in this region of the DNA sequence are most likely due to the cleavage of complex 2–ODN II adducts on both strands. Since the conditions used favor a maximum of one Pt–ODN adduct per duplex, these cleavages on nonmodified ODN I probably originate from complex 2–ODN II adducts.
The results are summarized in Fig. 9. Importantly, the cleavage of ODN I by Pt–ODN II adducts appears to be partially responsible of the visible nonselective cleavage of the duplex by complex 2.
Interestingly, the weak peaks observed on the 5′-side of the cleavage fragments (and shown with an asterisk in Fig. 8) probably correspond to DNA fragments with 3′-phosphoglycolate extremities. Indeed, these species are observed at the position of the cleavage products of the duplex produced by the action of complex 3, and which have been previously characterized [15]. Such fragments were earlier identified as the result of the oxidation of the C4′ position of 2-deoxyribose. Further investigations are required to confirm these proposals.
Concluding remarks
The novel heterodinuclear platinum–copper complex 2 exhibits improved nuclease activity, compared with its parent compound Cu(3-Clip-Phen) (3), and is able to perform double-strand breaks. The platinum moiety acts as an anchor to DNA, forcing the Cu(3-Clip-Phen) group to generate cuts in close proximity to the Pt–DNA adducts, hence even allowing direct double-strand breaks. Mechanistic investigations on a 36-bp DNA fragment (ODN I–ODN II) revealed that platinum adducts are indeed formed with both complexes 1 and 2. The platinum moiety of complexes 1 and 2 binds to GG (primarily) and AG sites, like cisplatin. Nevertheless, the Taq polymerase enzyme stops at different base pairs for cisplatin and complexes 1 and 2. This feature suggests that complexes 1 and 2 induce different distortions (compared with cisplatin) upon their binding to DNA, most likely owing to the bulkiness of the 3-Clip-Phen moiety. Complex 1 with added copper (1 equiv) shows a sequence-selective cleavage, in the close proximity of the platinum adducts. A sequence-selective cleavage is also observed for complex 2, but to a lesser extent compared with complex 1. The opposite strand also contains Pt–DNA adducts, but no clear sequence selectivity is observed, most likely owing to the lack of preferential platinum binding sites. These atypical platinum adducts lead to the partial damage of the other DNA strand, as is clearly evidenced by comparison of the cleavage products on a single-stranded or duplex DNA.
Interestingly, it has been found that piperidine is also able to activate the Cu(3-Clip-Phen) component of the bifunctional platinum–copper complexes, a feature so far not reported. Thus, a good sequence-selective cleavage is observed upon treatment of complex 1 with piperidine. Reasonable sequence selectivity is achieved with complex 2. The investigation of the mechanism of cleavage by complex 2 is currently in progress.
Abbreviations
- 3-Clip-Phen:
-
1-(1,10-Phenanthrolin-3-yloxy)-3-(1,10-phenanthrolin-8-yloxy)propan-2-amine
- DCM:
-
Dichloromethane
- DMF:
-
Dimethylformamide
- DMSO:
-
Dimethyl sulfoxide
- ESI:
-
Electrospray ionization
- HEPES:
-
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- MS:
-
Mass spectrometry
- ODN:
-
Oligonucleotide
- TBE:
-
2-Amino-2-(hydroxymethyl)propane-1,3-diol borate edta
- Tris:
-
2-Amino-2-(hydroxymethyl)propane-1,3-diol
References
Chabner BA, Roberts TG (2005) Nat Rev Cancer 5:65–72
Hurley LH (2002) Nat Rev Cancer 2:188–200
Reedijk J (2003) Proc Natl Acad Sci USA 100:3611–3616
Chen JY, Stubbe J (2005) Nat Rev Cancer 5:102–112
Umezawa H, Maeda K, Takeuchi T, Okami Y (1966) J Antibiot 19:200–209
Burrows CJ, Muller JG (1998) Chem Rev 98:1109–1151
Pitié M, Boldron C, Pratviel G (2006) DNA oxidation by copper and manganese complexes. Elsevier Academic Press, San Diego
Pratviel G, Bernadou J, Meunier B (1995) Angew Chem Int Ed Engl 34:746–769
Sigman DS, Mazumder A, Perrin DM (1993) Chem Rev 93:2295–2316
Marshall LE, Graham DR, Reich KA, Sigman DS (1981) Biochemistry 20:244–250
Veal JM, Rill RL (1991) Biochemistry 30:1132–1140
Zelenko O, Gallagher J, Sigman DS (1997) Angew Chem Int Ed Engl 36:2776–2778
Goyne TE, Sigman DS (1987) J Am Chem Soc 109:2846–2848
Kuwabara M, Yoon C, Goyne T, Thederahn T, Sigman DS (1986) Biochemistry 25:7401–7408
Pitié M, Sudres B, Meunier B (1998) Chem Commun 2597–2598
Boldron C, Ross SA, Pitié M, Meunier B (2002) Bioconjug Chem 13:1013–1020
Pitié M, Van Horn JD, Brion D, Burrows CJ, Meunier B (2000) Bioconjug Chem 11:892–900
Pitié M, Burrows CJ, Meunier B (2000) Nucleic Acids Res 28:4856–4864
de Hoog P, Boldron C, Gamez P, Sliedregt-Bol K, Roland I, Pitié M et al (2007) J Med Chem 50:3148–3152
Rosenberg B, van Camp L, Trosko JE, Mansour VH (1969) Nature 222:385–386
Jamieson ER, Lippard SJ (1999) Chem Rev 99:2467–2498
Kostrhunova H, Brabec V (2000) Biochemistry 39:12639–12649
Robillard MS, Davies NP, van der Marel GA, van Boom JH, Reedijk J, Murray V (2003) J Inorg Biochem 96:331–338
Temple MD, McFadyen WD, Holmes RJ, Denny WA, Murray V (2000) Biochemistry 39:5593–5599
Fasman G (1975) Handbook of biochemistry and molecular biology: nucleic acids. CRC Press, Boca Raton, p 175
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Maxam AM, Gilbert W (1977) Proc Natl Acad Sci USA 74:560–564
Haber JE (2000) Trends Genet 16:259–264
Karran P (2000) Curr Opin Genet Dev 10:144–150
Ackley MC, Barry CG, Mounce AM, Farmer MC, Springer BE, Day CS et al (2004) J Biol Inorg Chem 9:453–461
Bassett E, Vaisman A, Havener JM, Masutani C, Hanaoka F, Chaney SG (2003) Biochemistry 42:14197–14206
Galea AM, Murray V (2002) Biochim Biophys Acta Gene Struct Expr 1579:142–152
Kasparkova J, Novakova O, Farrell N, Brabec V (2003) Biochemistry 42:792–800
Murray V, Motyka H, England PR, Wickham G, Lee HH, Denny WA et al (1992) Biochemistry 31:11812–11817
Murray V, Motyka H, England PR, Wickham G, Lee HH, Denny WA et al (1992) J Biol Chem 267:18805–18809
Temple MD, Recabarren P, McFadyen WD, Holmes RJ, Denny WA, Murray V (2002) Biochim Biophys Acta Gene Struct Expr 1574:223–230
Vaisman A, Chaney SG (2000) J Biol Chem 275:13017–13025
Woynarowski JM, Chapman WG, Napier C, Herzig MCS, Juniewicz P (1998) Mol Pharmacol 54:770–777
Zou Y, Vanhouten B, Farrell N (1994) Biochemistry 33:5404–5410
Petitjean A, Barton JK (2004) J Am Chem Soc 126:14728–14729
Thurston DE, Morris SJ, Hartley JA (1996) Chem Commun 563–565
Acknowledgments
The authors are indebted to CERC3, CNRS France and the Chemical Research Council of the Netherlands, for financial support. The authors wish to thank Johnson & Matthey (Reading, UK) for their generous loan of K2PtCl4.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
de Hoog, P., Pitié, M., Amadei, G. et al. DNA cleavage and binding selectivity of a heterodinuclear Pt–Cu(3-Clip-Phen) complex. J Biol Inorg Chem 13, 575–586 (2008). https://doi.org/10.1007/s00775-008-0346-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-008-0346-y