Abstract
Numerous observations from recombinant systems have shown that properties such as the specific cell growth rate and the plasmid-free cell formation rate are related, not only to the average plasmid content per cell, but also to the plasmid distribution within a population. The plasmid distribution in recombinant cultures can have an effect on the culture productivity that cannot be modelled using average values of the overall culture. The prediction of the behaviour of a plasmid content distribution and its causes and effects can only be studied using segregated models. A segregated model that describes populations of recombinant cells characterized by their plasmid content distribution has been developed. This model includes critical causes of recombinant culture instability such as the plasmid partition mechanism at cell division, plasmid replication kinetics and the effect of the plasmid content on the specific growth rate. The segregated model allows investigation of the effect of each of these causes and that of the plasmid content distribution on the observable behaviour of a recombinant culture.
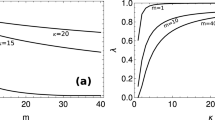
The effect of two partitioning mechanisms (Gaussian distribution and binomial distribution) on culture stability was investigated. The Gaussian distribution is slightly more stable. A small plasmid replication rate constant results in a very unstable culture even after short periods of time. This instability is dramatically improved for a larger value of this constant, hence improving protein synthesis. For a very narrow initial plasmid distribution, a given plasmid replication rate and partitioning mechanism can become broad even after a relatively short period of time. In contrast, a very "broad" initial distribution gave rise to a "Gamma-like" distribution profile. If we compare the results obtained in the simulations of the segregated model with those of the non-segregated one (average model), the latter model predicts much more stable behaviour, thus these average models cannot predict culture instability with the same precision.
When compared with the experimental results, the segregated model was able to predict the practical behaviour with accuracy even in a system with a high plasmid content per cell and a high rate of plasmid-free cell formation which could not be achieved with a non-segregated model.







Similar content being viewed by others
Abbreviations
- A :
-
dimensionless number, defined in Table 2
- D :
-
dilution rate, 1/h
- E :
-
recombinant enzyme concentration, U/L
- f(z,t):
-
probability density function
- F :
-
fraction of plasmid-containing cells
- H :
-
function defined in Table 4
- h :
-
ratio Δt*/Δz*
- I:
-
number of nodes in the z variable
- k E :
-
recombinant enzyme rate constant, [g/(plasmid/cell)(cell/L)h]
- K Eμ :
-
inhibition constant in Eq. (14), 1/h
- K S :
-
Monod constant, g/L
- K z :
-
inhibition constant in Eq. (7), (plasmid/cell)n
- K zz :
-
plasmid replication rate constant Eq. (8), 1/h
- K zμ :
- m :
-
power in Eq. (7)
- n(t):
-
t)cell concentration, cell/L
- p(z,z′):
-
partitioning probability function
- r z :
-
plasmid replication kinetics, plasmid/h cell
- R :
-
function defined in Table 4
- S :
-
substrate concentration, g/L
- t :
-
time, h
- V 0 :
-
plasmid replication rate constant, Eq. (8), plasmid/cell h
- V :
-
discretized v(z,t*) distribution function
- w(z,t):
-
distribution of plasmid-containing cells
- Y wS :
-
substrate yield coefficient, g/g
- Z :
-
space of possible values of variable z
- α,β :
-
parameters in Eq. (12)
- Δt :
-
time step size, h
- Δz :
-
plasmid content step size, plasmid/cell
- ε :
-
standard deviation
- Γ :
-
gamma function
- μ :
-
specific growth rate, 1/h
- θ :
-
probability of formation a plasmid-free cell
- *:
-
dimensionless variables
- av:
-
average
- max:
-
maximum
- F:
-
feed
- 0:
-
initial
References
Summers DK, Sherratt DJ (1984) Cell 36:1097–1103
Leonhardt H., Alonso JC (1991) Gene 103:107–111
Koizumi JI, Monden Y, Aiba S (1985) Biotechnol. Bioeng. 27:721–728
Hopkins DJ, Betenbaugh MJ, Dhurjati P. (1987) Biotechnol Bioeng 29:85–91
Ramírez DM, Bentley WE (1993) Biotechnol Bioeng 41:557–565
McLoughlin AJ (1994) Biotech Adv 12:279–324
Lee SB, Bailey JE (1984) Biotechnol Bioeng 26:66–73
Lin Chao S, Bremer H (1986) Mol Genet 203:143–149
Shene C, Mir N, Andrews B, Asenjo JA (1996) Ann NY Acad Sci 782:334–349
Fredrickson AG, Ramkrishna D, Tsuchiya HM (1967) Math Biosci 1:327–374
Eakman JM, Fredrickson AG, Tsuchiya HM (1966) Chem Eng Prog Symp Ser 69, 62:37–49
Ramkrishna D (1992) Adv Biochem Eng Biotech 46:1–47
Subramarian G, Ramkrishna D (1971) Math Biosci 10:1–23
Hunter JB, Asenjo JA (1990) Biotechnol Bioeng 35:31–42
Hardjito L, Greenfield PF, Lee PL (1992) Biotechnol Prog 8:298–306
Lee SB, Ryu DDY, Seigel R, Park SH (1988) Biotechnol Bioeng 31:805–820
Park SH, Ryu DDY, Lee SB (1991) Biotechnol Bioeng 37:404–414
Satyagal VN, Agrawal P (1989) Biotechnol Bioeng 33:1135–1144
Togna AP, Fu J, Shuler ML (1993) Biotechnol Bioeng 42:557–570
Wei D, Parulekar SJ, Stark BC, Weigand WA (1989) Biotechnol Bioeng 33:1010–1020
Bentley WE Quiroga OE (1993) Biotechnol Bioeng 42:222–234
Kim BG, Shuler ML (1990) Biotechnol Bioeng 36:581–592
Ataii MM, Shuler ML (1987) Biotechnol Bioeng 30:389–397
Seo JH, Bailey JE (1985b) Biotechnol Bioeng 27:1668–1674
Kothari IR, Martin GC, Reilly PJ, Martin PJ, Eakman JM (1972) Biotechnol Bioeng 14:915–938
Mitchell AR, Griffiths DF (1980) The finite difference method in partial differential equations. John Wiley
Leonhardt H. (1990) Gene 94:121–124
Acknowledgments
This research was supported by Conicyt (project Fondecyt 1950620) and the Millennium Institute for Advanced Studies in Cell Biology and Biotechnology (ICM P 99-031-F).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shene, C., Andrews, B.A. & Asenjo, J.A. Study of recombinant micro-organism populations characterized by their plasmid content per cell using a segregated model. Bioprocess Biosyst Eng 25, 333–340 (2003). https://doi.org/10.1007/s00449-002-0313-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-002-0313-x