Abstract
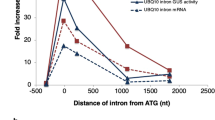
Introns play a very important role in regulating gene expression in eukaryotes. In plants, many introns enhance gene expression, and the effect of intron-mediated enhancement (IME) of gene expression is reportedly often more profound in monocots than in dicots. To further gain insight of IME in monocot plants, we quantitatively dissected the effect of the 5′ UTR intron of the rice rubi3 gene at various gene expression levels in stably transformed suspension cell lines. The intron enhanced the GUS reporter gene activity in these lines by about 29-fold. Nuclear run-on experiments demonstrated a nearly twofold enhancement by the 5′ UTR intron at the transcriptional level. RNA analysis by RealTime quantitative RT-PCR assays indicated the intron enhanced the steady state RNA level of the GUS reporter gene by nearly 20-fold, implying a strong role of the intron in RNA processing and/or export. The results also implicated a moderate role of the intron in enhancement at the translational level (∼45%). Moreover, results from a transient assay experiment using a shortened exon 1 sequence revealed an important role of exon 1 of rubi3 in gene expression. It may also hint a divergence in IME mechanisms between plant and animal cells. These results demonstrated transcriptional enhancement by a plant intron, but suggested that post-transcriptional event(s) be the major source of IME.




Similar content being viewed by others
References
Azhakanandam K, McCabe MS, Power JB, Lowe KC, Cocking EC, Davey MR (2000) T-DNA transfer, integration, expression and inheritance in rice: effects of plant genotype and Agrobacterium super-virulence. J Plant Physiol 157:429–439
Belostotsky DA, Rose AB (2005) Plant gene expression in the age of systems biology: integrating transcriptional and post-transcriptional events. Trends Plant Sci 10:347–353
Blockland RV, Geest NV, Mol JNM, Kooter JM (1994) Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase RNA turnover. Plant J 6:861–877
Bourdon V, Harvey A, Lonsdale DM (2001) Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep 2:394–398
Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765–775
Buchman AR, Berg P (1988) Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol 8:4395–4405
Callis J, Fromm ME, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1:1183–1200
Chekanova JA, Dutko JA, Mian IS, Belostotsky DA (2002) Arabidopsis thaliana exosome subuint AtRrp4p is a hydrolitic 3′→5′ exonuclease containing S1 and KH RNA binding domains. Nucleic Acids Res 30:695–700
Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218
Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7:120
Clancy M, Hannah LC (2002) Splicing of the maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol 130:918–929
Clancy M, Vasil V, Hannah LC, Vasil IK (1994) Maize Shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Sci 98:151–161
Dean C, Favreau M, Bondnutter D, Bedbrook J, Dunsmuir P (1989) Sequences downstream of translation start regulate quantitative expression of 2 Petunia rbcs genes. Plant Cell 1:201–208
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 14:19–21
Egoavil C, Marton HA, Baynton CE, McCullough AJ, Schuler MA (1997) Structural analysis of elements contributing to 5′ splice site selection in plant pre-mRNA transcripts. Plant J 12:971–980
Eick D, Kohlhuber LF, Wolf DA, Strobl LJ (1994) Activation of pausing RNA polymerases by nuclear run-on experiments. Anal Biochem 218:347–351
Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ (2002) Promoter proximal splice sites enhance transcription. Genes Dev 16:2792–2799
Gallagher SR (1992) GUS protocols: using the GUS gene as a reporter of gene expression quantitation of GUS activity by fluorometry. Academic press Inc., New York
Gallie DR (1993) Posttranscriptional regulation of gene-expression in plants. Annu Rev Plant Physiol Plant Mol Biol 44:77–105
Goodall GJ, Filipowicz W (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58:473–483
Goodall GJ, Kiss T, Filipowicz W (1991) Nuclear RNA splicing and small nuclear RNAs and their genes in higher plants. Oxf Surv Plant Mol Cell Biol 7:255–296
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Huang MTF, Gorman CM (1990) Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res 18:937–947
Ingelbrecht I, de Carvalho F (1992) Isolation of nuclei and in vivo run-off transcription. In: Inze D, Van der Straeten D, Van Montagu M (eds) EMBO practical course on plant molecular biology. Laboratorium voor Genetica, Gent, pp 117–132
Ingham DJ, Beer S, Money S, Hansen G (2001) Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31:132–140
Initiative AG (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Jefferson RA, Kavanagh TA, Bevan MW (1987) Gus fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G (2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6:673–682
Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G (2001) Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J 20:6424–6433
Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25:1869–1872
Klein TM, Gradziel T, Fromm ME, Sanford JC (1988) Factors influencing gene delivery into Zea mays cells by high-velocity microprojectiles. Biotechnology 6:559–563
Lambermon MHL, Simpson GG, Kirk DAW, Hemmings-Mieszczak M, Klahre U, Filipowicz W (2000) UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J 19:1638–1649
Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome depositsmultiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19:6860–6869
Lu J (2006) Gene expression regulation by the 5′ regulatory sequences of the rice rubi3 gene in transgenic rice plants. Ph.D. thesis. North Carolina State University. http://www.lib.ncsu.edu/theses/available/etd-04272006-145216/unrestricted/etd.pdf
Lu SH, Cullen BR (2003) Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9:618–630
Luehrsen KR, Walbot V (1991) Intron enhancement of gene-expression and the splicing efficiency of introns in maize cells. Mol Gen Genet 225:81–93
Luehrsen KR, Walbot V (1994) Addition of A-rich and U-rich sequence increases the splicing efficiency of a deleted form of a maize intron. Plant Mol Biol 24:449–463
Luehrsen KR, Taha S, Walbot V (1994) Nuclear pre-messenger RNA processing in higher plants. Nucleic Acid Res 47:149–193
Mascarenhas D, Mettler IJ, Pierce DA, Lowe HW (1990) Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol 15:913–920
McCullough AJ, Schuler MA (1997) Intronic and exonic sequences modulate 5’ splice site selection in plant nuclei. Nucleic Acids Res 25:1071–1077
McElroy D, Zhang WG, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2:163–171
Nott A, Muslin SH, Moore MJ (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9:607–617
Nott A, Le Hir H, Moore MJ (2004) Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Gene Dev 18:210–222
Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A (1993) Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135:385–404
Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16:260–269
Proudfoot NJ, Furger A, Dye MJ (2002) Integrating rnRNA processing with transcription. Cell 108:501–512
Reddy ASN (2001) Nuclear pre-mRNA splicing in plants. Crit Rev Plant Sci 20:523–571
Rethmeier N, Seurinck J, VanMontagu M, Cornelissen M (1997) Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J 12:895–899
Rethmeier N, Kramer E, van Montagu M, Cornelissen M (1998) Identification of cat sequences required for intron-dependent gene expression in maize cells. Plant J 13:833–835
Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8:1444–1453
Rose AB (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40:744–751
Rose AB, Beliakoff JA (2000) Intron-mediated enhancement of gene expression independent of unique intron sequence and splicing. Plant Physiol 122:535–542
Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11:455–464
Sanford JR, Gray NK, Beckmann K, Caceres JF (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18:755–768
Simpson GG, Filipowicz W (1996) Splicing of precursors to mRNA in higher plants: Mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol 32:1–41
Simpson CG, Simpson GG, Clark G, Leader DJ, Vaux P, Guerineau F, Waugh R, Brown JWS (1992) Splicing of plant pre-messenger RNAs. Proc R Soc Edinb B Biol Sci 99:31–50
Sivamani E, Qu R (2006) Expression enhancement of a rice polyubiquitin gene promoter. Plant Mol Biol 60:225–239
Sivamani E, Huet H, Shen P, Ong CA, de Kochko A, Fauquet C, Beachy RN (1999) Rice plant (Oryza sativa L.) containing Rice tungro spherical virus (RTSV) coat protein transgenes are resistant to virus infection. Mol Breed 5:177–185
Wiegand HL, Lu SH, Cullen BR (2003) Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA 100:11327–11332
Yon J, Fried M (1989) Precise gene fusion by PCR. Nucleic Acids Res 17:4895–4895
Zhang S, Song W, Chen L, Ruan D, Taylor N, Ronald P, Beachy RN, Fauquet CM (1998) Transgenic elite Indica rice varieties, resistant to Xanthomonas oryzae pv. Oryzae. Mol Breed 4:551–558
Acknowledgments
The project was, in part, supported by grants from the Consortium for Plant Biotechnology Research, Inc. (St. Simons Island, GA, USA, GO12026-162 and GO12026-171 to R.Q.) and Syngenta Biotechnology Inc. (Research Triangle Park, NC, USA). The authors thank Dr. J. Nicholson for technical advice on nuclear run-on assay, Drs. L. Mathies and F. Corbin for using their facilities, and M. Massel for technical assistance. We also like to thank Drs. R. Wu and M. Ryba-White for providing rice Act1 gene clones. The authors are grateful to Plant Analysis Group at Syngenta Biotechnology Inc. for performing the RealTime PCR assays to determine transgene copy number, and to Dr. R. Shi for technical suggestions in the RealTime qRT-PCR assays. Our gratitude also extends to Dr. R. S. Boston for helpful discussion and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Akhilesh Tyagi.
Rights and permissions
About this article
Cite this article
Samadder, P., Sivamani, E., Lu, J. et al. Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Genet Genomics 279, 429–439 (2008). https://doi.org/10.1007/s00438-008-0323-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0323-8