Abstract
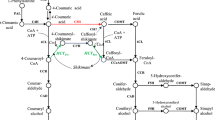
Flowering plants have syringyl and guaiacyl subunits in lignin in contrast to the guaiacyl lignin in gymnosperms. The biosynthesis of syringyl subunits is initiated by coniferaldehyde 5-hydroxylase (CAld5H). In Populus trichocarpa there are two closely related CAld5H enzymes (PtrCAld5H1 and PtrCAld5H2) associated with lignin biosynthesis during wood formation. We used yeast recombinant PtrCAld5H1 and PtrCAld5H2 proteins to carry out Michaelis–Menten and inhibition kinetics with LC–MS/MS based absolute protein quantification. CAld5H, a monooxygenase, requires a cytochrome P450 reductase (CPR) as an electron donor. We cloned and expressed three P. trichocarpa CPRs in yeast and show that all are active with both CAld5Hs. The kinetic analysis shows both CAld5Hs have essentially the same biochemical functions. When both CAld5Hs are coexpressed in the same yeast membranes, the resulting enzyme activities are additive, suggesting functional redundancy and independence of these two enzymes. Simulated reaction flux based on Michaelis–Menten kinetics and inhibition kinetics confirmed the redundancy and independence. Subcellular localization of both CAld5Hs as sGFP fusion proteins expressed in P. trichocarpa differentiating xylem protoplasts indicate that they are endoplasmic reticulum resident proteins. These results imply that during wood formation, 5-hydroxylation in monolignol biosynthesis of P. trichocarpa requires the combined metabolic flux of these two CAld5Hs to maintain adequate biosynthesis of syringyl lignin. The combination of genetic analysis, absolute protein quantitation-based enzyme kinetics, homologous CPR specificity, SNP characterization, and ER localization provides a more rigorous basis for a comprehensive systems understanding of 5-hydroxylation in lignin biosynthesis.









Similar content being viewed by others
Abbreviations
- SDX:
-
Stem differentiating xylem
- CPR:
-
Cytochrome P450 reductase
- CAld5H:
-
Coniferaldehyde 5-hydroxylase
- PC-IDMS:
-
Protein cleavage isotope dilution mass spectrometry
- ER:
-
Endoplasmic reticulum
References
Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC (2004) Absolute quantification of the model biomarker prostate-specific antigen in serum by LC–MS/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res 3:644–652
Barr JR, Maggio VL, Patterson DG Jr, Cooper GR, Henderson LO, Turner WE, Smith SJ, Hannon WH, Needham LL, Sampson EJ (1996) Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A–I. Clin Chem 42:1676–1682
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Brown SA (1961) Chemistry of lignification: biochemical research on lignins is yielding clues to the structure and formation of these complex polymers. Science 134:305–313
Brown SA, Neish AC (1955a) Studies of lignin biosynthesis using isotopic carbon. IV. Formation from some aromatic monomers. Can J Biochem Physiol 33:948–962
Brown SA, Neish AC (1955b) Shikimic acid as a precursor in lignin biosynthesis. Nature 175:688–689
Brown SA, Neish AC (1956) Studies of lignin biosynthesis using isotopic carbon. V. Comparative studies on different plant species. Can J Biochem Physiol 34:769–778
Brown SA, Neish AC (1959) Studies of lignin biosynthesis by use of isotopic carbon. VIII. Isolation of radioactive hydrogenolysis products of lignin. J Am Chem Soc 81:2419–2424
Chen HC, Shuford CM, Liu J, Muddiman DC, Sederoff RR, Chiang VL (2011) Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci 108:21253–21258
Dass C, Kusmierz JJ, Desiderio DM (1991) Mass spectrometric quantification of endogenous beta-endorphin. Biol Mass Spectrom 20:130–138
Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by Tandem MS. P Natl Acad Sci USA 100:6940–6945
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG. Method Nat Protocol 2:31–34
Grand C (1984) Ferulic acid 5-hydroxylase: a new cytochrome P-450-dependent enzyme from higher plant microsomes involved in lignin synthesis. FEBS Lett 169:7–11
Higuchi T (1997) Biochemistry and molecular biology of wood. Springer, New York, pp 131–233
Higuchi T, Brown SA (1963a) Studies of lignin biosynthesis using isotopic carbon. XII. The biosynthesis and metabolism of spinapic acid. Can J Biochem Physiol 41:613–620
Higuchi T, Brown SA (1963b) Studies of lignin biosynthesis using isotopic carbon: XIII. The phenylpropanoid system in lignification. Biochem Cell Biol 41:621–628
Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci 96:10045–10050
Jiang H, Morgan JA (2004) Optimization of an in vivo plant P450 monooxygenase system in Saccharomyces cerevisiae. Biotechnol Bioeng 85:130–137
Kushnir MM, Rockwood AL, Nelson GJ, Yue BF, Urry FM (2005) Assessing analytical specificity in quantitative analysis using tandem mass spectrometry. Clin Biochem 38:319–327
Lange V, Picotti P, Domon B, Aebersold R (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4:14
Maclean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, Maccoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968
Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci 96:8955–8960
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133(3):1051–1071
Ralph J (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3:29–60
Ralph J, Brunow G, Boerjan W (2007) Lignins. In: Encyclopedia of life sciences. Wiley, Chichester
Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J (2001) Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch Biochem Biophys 393:222–235
Rogers LA, Campbell MM (2004) The genetic control of lignin deposition during plant growth and development. New Phytol 164:17–30
Sarkanen KV, Ludwig CH (1971) Lignins: occurrence, formation, structure and reactions. Wiley, New York
Sauvage FL, Gaulier JM, Lachatre G, Marquet P (2008) Pitfalls and prevention strategies for liquid chromatography-tandem mass spectrometry in the selected reaction-monitoring mode for drug analysis. Clin Chem 54:1519–1527
Scopes RK (1974) Measurement of protein by spectrophotometry at 205 nm. Anal Biochem 59:277–282
Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL (2009) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51:144–163
Shuford CM, Li Q, Sun YH, Chen HC, Wang JP, Shi R, Sederoff RR, Chiang VL, Muddiman DC (2012) Comprehensive quantification of monolignol-pathway enzymes in Populus trichocarpa by protein cleavage isotope dilution mass spectrometry (submitted)
Tuskan GA et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana ADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272:19176–19186
Whetten R, Sederoff R (1995) Lignin biosynthesis. Plant Cell 7:1001–1003
Wright D, Brown SA, Neish AC (1958) Studies of lignin biosynthesis using isotopic carbon. VI. Formation of the side chain of the phenylpropane monomer. Can J Biochem Physiol 36:1037–1045
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocol 2:1565–1572
Acknowledgments
We thank Dr. Danièle Werck-Reichhart and Dr. Denis Pompon for the plasmid pYeDP60. This work was supported by National Science Foundation Plant Genome Research Program Grant DBI-0922391 (to V.L.C).
Author information
Authors and Affiliations
Corresponding author
Additional information
A contribution to the Special Issue on Metabolic Plant Biology.
Rights and permissions
About this article
Cite this article
Wang, J.P., Shuford, C.M., Li, Q. et al. Functional redundancy of the two 5-hydroxylases in monolignol biosynthesis of Populus trichocarpa: LC–MS/MS based protein quantification and metabolic flux analysis. Planta 236, 795–808 (2012). https://doi.org/10.1007/s00425-012-1663-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1663-5