Abstract
Sex chromosome genes control sex determination and differentiation, but the mechanisms of sex determination in birds are unknown. In this study, we analyzed the gene FEM1C which is highly conserved from Caenorhabditis elegans to higher vertebrates and interacts with the sex determining pathway in C. elegans. We found that FEM1C is located on the Z and W chromosome of zebra finches and probably other Passerine birds, but shows only Z linkage in other avian orders. In the zebra finch, FEM1C-W is degraded because of a point mutation and possibly because of loss of the first exon containing the start methionine. Thus, FEM1C-W appears to have degenerated or been lost from most bird species. FEM1C-Z is expressed in a cytoplasmic location in zebra finch fibroblast cells, as in C. elegans. FEM1C represents an interesting example of evolutionary degradation of a W chromosome gene.
Similar content being viewed by others
Introduction
In birds, the female sex chromosomes are ZW and male ZZ. The ancestral chromosomes of avian sex chromosomes are different from those of eutherian mammals and snakes (Fridolfsson et al. 1998; Nanda et al. 1999; Matsubara et al. 2006), although recent evidence indicates surprising shared gene content of the avian Z and monotreme X chromosomes (Rens et al. 2007). The gene content of the Z chromosome appears to have changed little during avian evolution, as shown by comparative painting analysis (Itoh and Arnold 2005; Nishida-Umehara et al. 2007; Tsuda et al. 2007), even in Ratites (Struthioniformes) which are considered to be an ancestral avian species whose sex chromosomes are morphologically relatively undifferentiated (Ogawa et al. 1998; Shetty et al. 1999). The sex determining system in birds is still unclear, although several candidate systems have been suggested. One possibility is that the W chromosome has a dominant ovary-determining gene, such as ASW/Wpkci/HINT1W (Hori et al. 2000; O’Neill et al. 2000) or FET-1 (Reed and Sinclair 2002). Another is that the Z chromosome contains a gene that causes development of testes because of a higher expressed dose of the gene in ZZ males relative to ZW females. One such candidate is the Z-linked testis differentiation gene, DMRT1, which might induce testis differentiation because of higher expression in the ZZ male compared to ZW female (Raymond et al. 1999). It is also possible that Z and W genes both contribute to gonadal differentiation (Graves 2003).
The sex determination of Caenorhabditis elegans is controlled by a signal transduction pathway including the fem-1 gene, which encodes an ankyrin repeat protein. Mutation of fem-1 leads to feminization (male to hermaphrodite sex reversal). The FEM-1, FEM-2, and FEM-3 proteins form a complex with CUL-2 that regulates the activity of the TRA-1, a DNA-binding zinc finger transcription factor that regulates the DM domain gene mab-3, which in turn regulates male sexual development (Sokol and Kuwabara 2000; Starostina et al. 2007; Kuwabara 2007).
The highly conserved Fem1 gene family in vertebrates comprises three genes Fem1a, Fem1b, and Fem1c (Ventura-Holman et al. 1998, 2003). Human FEM1C maps to chromosome 5q22, and the exon–intron structure and expression pattern in adult tissue are conserved between mouse and human (Ventura-Holman et al. 2003). Both human FEM1C and mouse Fem1c transcripts show alternative splicing (Ventura-Holman et al. 2003). The 6.2-kb mRNA is ubiquitously expressed in all mouse tissues, and additional 3.8- and 2.8-kb transcripts are present in mouse testis. Humans have a ubiquitously expressed 6.2- and 3.8-kb mRNA and a testis-specific 2.8-kb transcript. Schlamp et al. (2004) reported a slight non-significant increase in the number of female pups in a mouse line with reduced expression of Fem1c, caused by insertion of the βGeo gene trap vector in the first intron of Fem1c gene, and concluded that the gene had no known effect on sexual development or fertility.
According to classical comparative mapping analysis, the chicken Z chromosome has large blocks of sequences that are syntenic with human chromosomes 5, 8, 9, and 18 (Schmid et al. 2000). In this study, we analyzed the sequence of human chromosome 5 to identify new sex-linked genes in birds. FEM1C gene, homologous to a C. elegans sex-determining gene, was cloned as a new sex-linked gene in birds and its linkage was confirmed by Southern blot and fluorescent in situ hybridization (FISH). Interestingly, an avian zoo blot analysis indicated that the coding region of FEM1C gene is Z-linked in most birds but also has W linkage in Passerines. Further analysis of the sequence of BAC (bacterial artificial chromosome) DNA shows evolutionary degradation of FEM1C-W in zebra finch, which provides us with an interesting example of sex chromosome gene evolution.
Materials and methods
Southern blot hybridization and cloning of zebra finch FEM1C-Z cDNA
We identified the FEM1C gene as a candidate chicken Z-linked gene because it is present on human chromosome 5q in the region syntenic with chicken Z genes such as CHD1 or HINT1. The sex chromosome linkage of FEM1C was checked by Southern blot hybridization to male and female zebra finch genomic DNA as described (Itoh and Arnold 2005). Zebra finch total RNA was isolated from adult testis using Trizol Reagent (Invitrogen), and first-strand cDNA was synthesized by Superscript III reverse transcriptase (Invitrogen). Polymerase chain reaction (PCR) was carried out in a 25-μl mixture containing 0.2 mM each of dNTPs, 0.4 μM each of primers, 10 ng of cDNA as a template, 0.25 μl of 50× Titanium Taq polymerase (BD Biosciences), and one tenth volume of 10× Titanium Taq buffer (BD Biosciences). The primer sequences were designed to match highly conserved regions of human and mouse Fem1c sequences: FEM1C-3 (5′-GGG CAT ACT TGC TTG ATG AT-3′, 5′-AGC ATC TTC ATG ATG TCC A-3′) and FEM1C-4 (5′-TTG GAT ATG CAG CAG AGC AA-3′, 5′-ATA AGG AGA TTC ATG ATG TC-3′). The PCR reaction was carried out at 95°C for 4 min before the cycling reaction and 40 cycles of 95°C for 45 s/52°C for 30 s/72°C for 1 min and then followed by single cycle reaction at 72°C for 7 min. The PCR product was separated by gel electrophoresis. PCR products were cloned into pGEM-T Easy vector (Promega).
For cloning longer cDNAs, we used reverse transcription PCR (RT-PCR), 5′- or 3′-RACE or screened a female zebra finch post-hatch day 1 brain cDNA library. Z or W assignment of cDNA fragments was evaluated by the sequence comparison to Z- or W-derived BAC sequences. BAC clone 297J07 (for FEM1C-Z, Genbank AC192094) and 197F15 (for FEM1C-W, Genbank AC192093) were sequenced by the Washington University Genome Sequencing Center (http://www.genome.wustl.edu).
Sequence analysis
The sequences were edited by Vector NTI sequence analysis software. The open reading frames of C. elegans, human, mouse, zebra fish, chicken, and zebra finch sequences were aligned using Clustal W. Values of dN/dS comparing zebra finch FEM1C-Z and the predicted FEM1C-W were estimated by MEGA3.1 (Kumar et al. 2004). Phylogenetic trees were generated by MEGA3.1 using the neighbor-joining method. The map of dN/dS ratios along the gene sequence was calculated in 120-bp length segments (one tenth of the entire region) with overlapping windows of 60 nucleotides.
Northern blot hybridization
RNA of male and female adult zebra finch brain or gonads was isolated using Trizol (Invitrogen), then 8 μg/lane samples were subjected to denaturing agarose gel electrophoresis and capillary transfer to a MAGNA nylon transfer membrane (Osmonics) in 10× SSC. The membrane was hybridized with a 32P-labeled probe in the phosphate hybridization buffer (Itoh and Arnold 2005) at 68°C for 12 h then washed in 2× SSC, 0.1% sodium dodecyl sulfate (SDS) at room temperature for 5 min twice, 1× SSC, 0.1% SDS at 37°C for 15 min twice, and 65°C for 15 min twice and subjected to autoradiography.
Fluorescence in situ hybridization
Zebra finch BAC clones were isolated from a zebra finch BAC library made by the Arizona Genomics Institute (http://www.genome.arizona.edu; Luo et al. 2006). BAC clones were identified by probing membranes spotted with the zebra finch clones and further confirmed by probing Southern blots of the identified clones after restriction digest. BAC clone 297J07 (for FEM1C-Z) and 197F15 (for FEM1C-W) were used as probes in FISH to metaphase preparations of mitotic chromosomes prepared from zebra finch fibroblasts as described (Itoh and Arnold 2005).
GFP expression in fibroblasts
The coding region of FEM1C-Z was amplified by PCR and subcloned into pEGFP-N3 vector (Clontech). The zebra finch fibroblast cells were prepared from embryos and plated onto coverslips. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen) and cells were cultured for 24 h. After washing with 1× phosphate-buffered saline (PBS) twice, the cells were fixed with 4% paraformaldehyde /1× PBS at room temperature for 30 min. The nuclei were counterstained with DAPI and covered with VECTASHIELD mounting medium (Vector labs).
Results
Molecular cloning of zebra finch FEM1C gene
To search for novel avian ZW genes, we focused on genes encoded in a segment of human chromosome 5 homologous to chicken Z (Schmid et al. 2000), especially a block of genes including two known chicken chromosome Z genes, CHD1 and HINT1 (located at Hs 5q15-q21.1 and 5q23.2, respectively). In this segment, two genes drew our attention because of the expectation that Z genes might play special roles in sex determination or differentiation: FEM1C (5q22.3) and HSD17B4 (5q23.1). Zebra finch FEM1C cDNA fragments were amplified by RT-PCR using the primers matching a highly homologous region in human and mouse. HSD17B4 was from a zebra finch EST library (SB02032A1A09, Genbank CK313884). Southern blot analysis of male and female zebra finch genomic DNA using FEM1C and HSD17B4 cDNAs as probes suggested the existence of FEM1C-Z and -W sequences (FEM1C-4 probe; Fig. 1a), but no female-specific (W) hybridization of HSD17B4 W-form (data not shown). Interestingly, although the C-terminal coding region of FEM1C-Z (FEM1C-4, 1,095- to 1,644-bp region of FEM1C-Z: Genbank EU825191) detected both Z and W sequence, the N-terminal region of FEM1C (FEM1C-3, 445- to 650-bp region of FEM1C-Z) did not show any clear sign of W linkage (Fig. 1a, b). We then localized the FEM1C sequence on Z and W chromosome by FISH mapping using zebra finch BAC clones as a probe (Fig. 2). Because of the existence of ZBM repetitive sequence (Itoh et al. 2008), the BAC clones derived from FEM1C-Z and -W both stained the entire W chromosome. The physical position of FEM1C-Z was 38.8 ± 2.0% from p-terminal of Z chromosome.
Southern blot of hybridization genomic DNA, digested with HindIII or BamHI, from individual male (M) and female (F) of zebra finch probed with 32P-labeled zebra finch FEM1C-Z. The probes were from two different coding region of FEM1C-Z: a FEM1C-4, C-terminal and b FEM1C-3, N-terminal region. The existence of bands in a showing greater hybridization in males suggests Z linkage, and the female-specific bands suggest W linkage
Chromosome localization of BAC clone 297J07 encoding FEM1C-Z and 197F15 encoding FEM1C-W in zebra finch ZW female metaphase chromosome set. 297J07 BAC probe hybridized to both Z and W chromosomes because it includes repetitive sequences such as ZBM (Itoh et al. 2008). The position of FEM1C-Z on Z chromosome is 38.8 ± 2.0% from the p telomere. Scale bar, 10 μm
With RT-PCR, 5′- or 3′-RACE and screening for female zebra finch post-hatch day 1 brain cDNA library, we isolated a 3,108-bp zebra finch FEM1C-Z cDNA clone (Genbank EU825191). Despite considerable effort (RT-PCR using female RNA, screening of a female brain cDNA library, 5′- or 3′-RACE using several specific primers for FEM1C-W genomic sequence), we were not able to isolate a zebra finch FEM1C-W cDNA clone and therefore have no direct evidence for expression of FEM1C-W.
Characterization of zebra finch FEM1C-Z gene
We compared FEM1C gene amino acid sequences among chicken (XM_428816), zebra finch, human (NM_020177), mouse (NM_173423), zebra fish (NM_198145), and C. elegans (J03172; Fig. 3). The FEM1C amino acid sequence was highly conserved among C. elegans and higher vertebrates (consensus similarity: human–mouse, 99.7%; human–chicken, 94.5%; human–zebra fish, 86.1%; human–C. elegans, 46%; chicken–zebra finch, 96.8%), suggesting that it serves important functions. There were eight ankyrin repeat motif regions found by MOTIF search (http://motif.genome.jp/) for BLOCKS database (Fig. 3 and Table 1). Interestingly, vertebrates have three more ankyrin repeat sequences in the C-terminal half region relative to C. elegans. Assuming that the C elegans form of fem1c represents an ancestral form based on the single fem1 gene in that species, the addition of several whole ankyrin repeats in the lineage leading to the vertebrates suggests adaptive evolution of this region. The chicken does not have one of the ankyrin repeats, located at 482–494aa, as a result of one amino acid substitution.
Comparison of amino acid sequences of chicken (XM_428816), zebra finch (EU825191), human (NM_020177.2), mouse (NM_173423.3), zebra fish (NM_198145.2), and C. elegans (J03172) FEM1C. The positions of predicted ankyrin repeats (Table 2) are underlined
Southern blot hybridization of the FEM1C-4 probe, encoding the C-terminal region of FEM1C-Z, to an avian zoo blot detected both Z and W sequence in DNA of the common finch (Fig. 4a). Although the snowy owl, which is interestingly classified in the same “land birds” clade as zebra finch according to recent phylogenetic analysis (Hackett et al. 2008), had a very faint female-specific signal, other birds (except for ostrich) only had a Z-linked signal pattern. With the N-terminal region of FEM1C probe (FEM1C-3), no signal was detected in domestic duck and the signals in Japanese quail and snowy owl were faint (Fig. 4b), which would suggest poor conservation of this region in birds. The hybridization to ostrich was similar in the two sexes, suggesting that FEM1C is present in the large pseudoautosomal region common to the Z and W chromosome of that species, although minor divergence is not ruled out.
Zoo Southern blot analysis for Passerine (finch) and non-Passerine birds. Two FEM1C-Z probes were used: a probe FEM1C-4 encoding the C-terminal region which recognizes both Z and W in zebra finch, b probe FEM1C-3 encoding the N-terminal region which only recognizes Z in zebra finch. Four micrograms of HaeIII digested male or female genomic DNA was electrophoresed in each lane. Species include the chicken (Gallus g. domesticus), Japanese quail (Coturnix c. japonica), ostrich (Struthio camelus), king penguin (Aptenodytes patagonica), domestic duck (Anas plathynchos domestica), budgerigar (Melopsitacus undulatas), snowy owl (Nyctea scandiaca), and common finch (Lonchura striata var)
By northern blot analysis, the FEM1C is equally expressed in males and females (Fig. 5), but in gonads, the expression in testis is higher than in ovary. The ratio of FEM1C to GAPDH expression is not obviously different in testis vs. ovary, but the use of a loading control such as GAPDH is problematic because of the fundamental differences in cell types in the two tissues. It is not clear if the higher expression in testis suggests testis-specific function, particularly because most Z genes are not dosage-compensated and are thus expected to be expressed higher in males than females (Itoh et al. 2007; Ellegren et al. 2007; Arnold et al. 2008). The northern blot offers no strong evidence for alternative splicing in testis as occurs in mouse and human (Ventura-Holman et al. 2003).
To test FEM1C function further, we analyzed the cellular localization of green fluorescence protein (GFP) fusion protein for zebra finch FEM1C-Z. Interestingly, in zebra finch embryonic fibroblast cells, GFP-FEM1C-Z fusion protein was localized in cytoplasm (Fig. 6a, b), and this pattern was not observed in cells transfected with the EGFP protein expression as a control (Fig. 6c, d).
The localization of zebra finch FEM1C-Z-GFP fusion protein in the cell body (b). The nucleus of the same cell is stained with DAPI in a. As a control, the pEGFP-N3 vector was also transfected in zebra finch fibroblast (c, d). Unlike pEGFP-N3 vector control, FEM1C-Z-GFP fusion protein was excluded from nucleus, which suggests cytoplasmic localization of this protein
BAC sequence analysis of zebra finch FEM1C-W
We found no evidence for expression of a W form of FEM1C, but could have missed it if expression is restricted to a short developmental period or specific tissue. The absence of a W form in most bird species measured in the zoo blot (Fig. 4) supports the idea that a W form is not functionally important in birds. To clarify the sequence of FEM1C-W, we analyzed the sequences of two BAC clones encoding FEM1C-Z and -W. Unfortunately, the first one or two exons of FEM1C were missing in each clone. Converting these BAC-derived FEM1C sequences to amino acid sequences, the predicted FEM1C-W sequence aligned closely to that of FEM1C-Z, except for two stop codons detected inside the coding region (Fig. 7). Thus, at most, a truncated form of FEM1C-W is likely to be expressed.
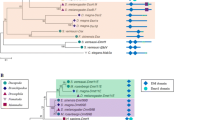
Table 2 shows the dN, dS, and dN/dS values for the comparison among human, mouse, chicken, and zebra finch. Higher dN/dS values denote either adaptive evolution or relaxed constraint. High conservation (low dN/dS) was observed between human and mouse, suggesting an important functional role for Fem1c in mammals. In contrast, the dN/dS value was markedly higher when comparing chicken vs. zebra finch not because of a lower dS which is about the same (~0.325) in both comparisons but because of a higher dN value (Table 2). This observation suggests higher conservation of Fem1c gene in mammals. In the comparison of zebra finch FEM1C-Z and -W, the dN/dS value was higher than chicken Z vs. zebra finch Z (Table 2) because of lower dS (0.271) and higher dN (0.036) relative to the chicken Z vs. zebra finch Z comparison (dS = 0.325, dN = 0.024). The phylogenetic tree of avian FEM1C genes (Fig. 8) suggested a closer relationship of zebra finch FEM1C-Z and -W than either with chicken FEM1C based on both nucleotide substitution and synonymous mutations (Fig. 8).
Neighbor-joining phylogenetic trees of the Fem1c gene family based on the currently available coding sequences. Branch lengths indicate relative relatedness. a p-distance for nucleotide substitutions resulting in any nucleotide substitutions. b p-distance for synonymous changes only. Bar, substitutions per nucleotide site
The divergence of the nucleotide sequences of FEM1C-Z and -W varies considerably along their lengths (Fig. 9). Although specific regions of the gene showed high or low dN/dS in the mammal–bird comparisons, there was no remarkable peak observed in the mammal to mammal (human to mouse) comparison (Fig. 9a). The pattern of peaks and valleys was quite similar when comparing mammal to either chicken or zebra finch (Fig. 9b). In bird to bird comparison (chicken to zebra finch), the same pattern was recognizable (Fig. 9c). These observations suggest FEM1C sequences in birds evolved mainly after divergence of birds and mammals. Moreover, evolutionary pressure was not equal along the sequence, but different bird species (chickens and zebra finches) show a common divergence from mammals, suggesting that the mammal–bird difference preceded the galliform–passerine split. Zebra finch FEM1C-Z and -W diversion was more significant compared to human–birds (Fig. 9d). The peaks occurred at three general positions, although the locations were slightly different from human–birds, one of which contains a stop codon. The other stop codon is outside the peak at amino acid position 195 in a region of evolutionary conservation (Figs. 7 and 9d). This pattern suggests that the nonsense mutations occur randomly relative to divergence on the Z and W sequences. There is no obvious relation between the positions of the valleys/peaks and ankyrin repeats.
The dN/dS values as a function of position along FEM1C genes. a The comparison of human–chicken (mammal–bird, black) and human–mouse (mammal–mammal, gray). FEM1C gene is highly conserved along the entire open reading frame between human and mouse. b Mammal–bird comparison: human–chicken Z (open square, solid line) and human–zebra finch Z (closed square, dashed line). The positional dN/dS values of chicken and zebra finch have similar pattern along the entire sequence. c Bird–bird comparison: chicken Z vs. zebra finch Z. The histograms show comparison of the two birds, and the line graphs of the mammal–bird comparison (b) are superimposed as a reference. The valleys and peaks of bird–bird dN/dS values followed the pattern of mammal–bird values, suggesting the region-specific changes occurred after divergence of mammals and birds. d The comparison of FEM1C-Z and predicted FEM1C-W in zebra finch. The graph of mammal–bird comparison (b) is shown as a reference. The valleys and peaks of zebra finch FEM1C-Z and -W have a pattern roughly similar to that of the mammal–bird comparison, but the values are larger. Asterisks are the positions of stop codons in FEM1C-W and open stars are the ankyrin repeat sites. The numbers on the abscissa are the amino acid positions on the zebra finch FEM1C-Z with the start Met as position 1
The zebra finch repetitive sequence ZBM (Itoh et al. 2008) was found in the FEM1C-W BAC clones. This ZBM repeat sequence is found at high density on the W chromosome but at lower density on the other chromosomes and tends to avoid gene coding regions (Itoh et al. 2008). The ZBM repetitive sequence is inserted into in 3′-UTR region of the predicted FEM1C-W sequence (Fig. 10).
Comparison of FEM1C-Z mRNA sequence and Z BAC 297J07 (a) and W BAC 197F15 (b). The sequences homologous to FEM1C-Z mRNA sequence are shown as a thick solid line in both Z and W BAC sequences and non-homologous genomic sequences as a dashed line. In both 297J07 and 197F15 BAC sequences, at least one exon covering the region from the start Met site to 543 bp is missing. In the Z chromosome BAC sequence, there was no intron sequence found in the region corresponding to the 544- to 3,108-bp region of FEM1C-Z cDNA sequence (Genbank: EU825191). The zebra finch W chromosome repetitive ZBM repeat sequence (Itoh et al. 2008) was found in the predicted 3’-UTR of FEM1C-W
Discussion
We have identified a FEM1C gene homologue in chicken by comparative analysis of the human genome database and confirmed its sex chromosome linkage in zebra finch. The FEM1C sequences were located on Z and W chromosome in snowy owl, common finch, and zebra finch, but only on the Z chromosome in several other non-ratite bird species. The FEM1C-W sequence in zebra finch contained two stop codons in the sequence we analyzed. Together with the absence of W sequences in other bird species, the insertion of stop codons in the zebra finch W sequence suggests that FEM1C-W gene in birds has or is undergoing degradation. If that is the case, however, it is surprising that much of the protein sequence of FEM1C-W is conserved in zebra finch. If the W form is totally non-functional because of the presence of stop codons, one would have expected the accumulation of other non-synonymous mutations in the coding sequence. Similar W degradation has been reported in ATP5A1 gene in parrots, but unlike FEM1C-W, the majority of species possesses a highly conserved ATP5A1W gene (de Kloet 2001). C. elegans fem-1 genes are critical for male sex determination; thus, if the avian FEM1C gene is also involved in a similar signaling cascade, the elimination of FEM1C-W might be advantageous to females. We did not detect any sex difference in expression of FEM1C-Z in brain, but the testis appeared to have slightly higher expression than the ovary. The cytoplasmic localization of FEM1C-Z gene in zebra finch fibroblast is similar to that of the C. elegans fem-1 gene. The presence of FEM1C on the Z chromosome is compatible with its potential role as a male-benefit testis gene (Vallender and Lahn 2004), but our results do not establish a function for FEM1C.
Birds diverged from mammals about 310 Mya (million years ago), and the monotreme platypus diverged from the other mammals about 210 Mya. In platypus, there are five X and five Y sex chromosomes, both of which form chains during the meiosis. One of the candidate avian sex determination genes, DMRT1, is localized on chromosome X5 in platypus. In human, the FEM1C gene is localized on human 5q22.3, close to the DMXL1 locus (5q23.1; Rens et al. 2007). FEM1C and DMXL1 have remained in a syntenic block during mammalian and avian evolution. The DMXL1 gene is on platypus chromosome X5 near DMRT1 (Rens et al. 2007), but FEM1C is not found in the platypus genome database (http://www.ensembl.org). Although both DMRT1 and FEM1C relate to the sex determination in fly and nematode, their role in birds and platypus are unknown even though they are situated on the sex chromosomes where a testis-determining role could be favored. The other Fem1 homologous genes, Fem1a and Fem1b, are found on chicken chromosome 28 and 10, respectively, and other components of the C. elegans sex-determining pathway (e.g., tra-1, fem-2, elc-1, and cul-2) show no conservation in the chicken genome so far.
Avian ZW chromosome genes reported so far appear to be conserved widely across species: SMAD2 (Mizuno et al. 2002), ATP5A1 (Nanda et al. 2000), UBE2R2 (Scholz et al. 2006), UBAP2 (Handley et al. 2004), NIPBL (Berlin and Ellegren 2006), SPIN (Itoh et al. 2001), HINT1 (Hori et al. 2000; O’Neill et al. 2000), CHD1 (Ellegren 1996). ZW genes are clustered on the chicken Z chromosome at the terminal end of short arm and the centromeric region of long arm (Fig. 11). FEM1C was located nearly at the terminal end of long arm in chicken genome database near what we believe to be the boundary of the Zq terminal heterochromatic region (Hori et al. 1996). The FEM1C-Z gene is therefore located outside of the other clusters of ZW genes. Based on reported evolutionary strata of Z chromosome using the divergence ratio between Z and W gene (Handley et al. 2004), the terminal region of short arm (including ATP5A1) differentiated about 58–85 Mya, and the centromeric region of Zq that includes SPIN, HINT1, and CHD1 is older and ceased recombination about 102–170 Mya. However, as we mentioned above, the recent chicken Z chromosome map suggests that ZW genes are restricted to specific regions although there is a large area unanalyzed because of lack of information on ZW genes. It is also possible that this region lacks ZW genes. At the terminal region of long arm where FEM1C-Z gene located, the homologous W sequence has completely diverged from Z in chicken, but this divergence has not been completed in zebra finch. These species comparisons suggest that it will be useful to examine the sequence of other species Z chromosome to analyze evolutionary strata and estimate the degree of ZW divergence.
The chicken FEM1C-Z gene is localized around the long arm terminal heterochromatic region of Z chromosome. The gene positions are based on chicken genome assembly 2 (May 2006, www.ensembl.com). The other ZW genes are clustered on the terminal region of short arm and the centromeric region of the long arm. The Zq heterochromatin was identified by Hori et al (1996), but is apparently absent from the current chicken Z sequence. Thus, the length of Z chromosome shown here is larger than the published chicken Z chromosome sequence
References
Arnold AP, Itoh Y, Melamed E (2008) A birds-eye view of sex chromosome dosage compensation. Annu Rev Genomics Hum Genet 9:109–127
Berlin S, Ellegren H (2006) Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J Mol Evol 62(1):66–72
de Kloet SR (2001) Loss of the gene for the alpha subunit of ATP synthase (ATP5A1) from the W chromosome in the African grey parrot (Psittacus erithacus). J Mol Evol 53(2):135–143
Ellegren H (1996) First gene on the avian W chromosome (CHD) provides a tag for universal sexing of non-ratite birds. Proc Biol Sci 263(1377):1635–1641
Ellegren H, Hultin-Rosenberg L, Brunström B, Dencker L, Kultima K, Scholz B (2007) Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol 5:40
Fridolfsson AK, Cheng H, Copeland NG, Jenkins NA, Liu HC, Raudsepp T, Woodage T, Chowdhary B, Halverson J, Ellegren H (1998) Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci U S A 95(14):8147–8152
Graves JA (2003) Sex and death in birds: a model of dosage compensation that predicts lethality of sex chromosome aneuploids. Cytogenet Genome Res 101(3–4):278–282
Hackett SJ, Kimball RT, Reddy S, Bowie RC, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T (2008) A phylogenomic study of birds reveals their evolutionary history. Science 320(5884):1763–1768
Handley LJ, Ceplitis H, Ellegren H (2004) Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167(1):367–376
Hori T, Suzuki Y, Solovei I, Saitoh Y, Hutchison N, Ikeda JE, Macgregor H, Mizuno S (1996) Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Res 4(6):411–426
Hori T, Asakawa S, Itoh Y, Shimizu N, Mizuno S (2000) Wpkci, encoding an altered form of PKCI, is conserved widely on the avian W chromosome and expressed in early female embryos: implication of its role in female sex determination. Mol Biol Cell 11(10):3645–3660
Itoh Y, Arnold AP (2005) Chromosomal polymorphism and comparative painting analysis in the zebra finch. Chromosome Res 13:47–56
Itoh Y, Hori T, Saitoh H, Mizuno S (2001) Chicken spindlin genes on W and Z chromosomes: transcriptional expression of both genes and dynamic behavior of spindlin in interphase and mitotic cells. Chromosome Res 9:283–299
Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP (2007) Dosage compensation is less effective in birds than in mammals. J Biol 6(1):2
Itoh Y, Kampf K, Arnold AP (2008) Molecular cloning of zebra finch W chromosome repetitive sequences: evolution of the avian W chromosome. Chromosoma 117(2):111–121
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5(2):150–163
Kuwabara PE (2007) A complex solution to a sexual dilemma. Dev Cell 13(1):6–8
Luo M, Yu Y, Kim H, Kudrna D, Itoh Y, Agate RJ, Melamed E, Goicoechea JL, Talag J, Mueller C, Wang W, Currie J, Sisneros NB, Wing RA, Arnold AP (2006) Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region. Genomics 87(1):181–190
Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y (2006) Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A 103(48):18190–18195
Mizuno S, Kunita R, Nakabayashi O, Kuroda Y, Arai N, Harata M, Ogawa A, Itoh Y, Teranishi M, Hori T (2002) Z and W chromosomes of chickens: studies on their gene functions in sex determination and sex differentiation. Cytogenet Genome Res 99(1–4):236–244
Nanda I, Shan Z, Schart M, Burt DW, Koehler M, Nothwang H, Grutzner F, Paton IR, Windsor D, Dunn I, Engel W, Staeheli P, Mizuno S, Haaf T, Schmid M (1999) 300 million years of conserved synteny between chicken Z and human chromosome 9. Nature Genet 21:258–259
Nanda I, Zend-Ajusch E, Shan Z, Grutzner F, Schart M, Burt DW, Koehler M, Fowler VM, Goodwin G, Schneider WJ, Mizuno S, Dechant G, Haaf T, Schmid M (2000) Conserved syntheny between the chicken Z sex chromosome and human chromosome 9 includes the male regulatory gene DMRT1: a comparative (re)view on avian sex determination. Cytogenet Cell Genet 89:67–78
Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y, Griffin DK (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res 15(6):721–734
Ogawa A, Murata K, Mizuno S (1998) The location of Z- and W-linked marker genes and sequence on the homomorphic sex chromosomes of the ostrich and the emu. Proc Natl Acad Sci U S A 95(8):4415–4418
O’Neill M, Binder M, Smith C, Andrews J, Reed K, Smith M, Millar C, Lambert D, Sinclair A (2000) ASW: a gene with conserved W-linkage and female specific expression in chick embryonic gonad. Dev Genes Evol 210(5):243–249
Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D (1999) Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol 215(2):208–220
Reed KJ, Sinclair AH (2002) FET-1: a novel W-linked, female specific gene up-regulated in the embryonic chicken ovary. Mech Dev 119(Suppl 1):S87–S90
Rens W, O’Brien PC, Grutzner F, Clarke O, Graphodatskaya D, Tsend-Ayush E, Trifonov VA, Skelton H, Wallis MC, Johnston S, Veyrunes F, Graves JA, Ferguson-Smith MA (2007) The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol 8(11):R243
Schlamp CL, Thliveris AT, Li Y, Kohl LP, Knop C, Dietz JA, Larsen IV, Imesch P, Pinto LH, Nickells RW (2004) Insertion of the beta Geo promoter trap into the FEM1C gene of ROSA3 mice. Mol Cell Biol 24(9):3794–3803
Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M, Schartl M, Haaf T, Weigend S, Fries R, Buerstedde JM, Wimmers K, Burt DW, Smith J, A’Hara S, Law A, Griffin DK, Bumstead N, Kaufman J, Thomson PA, Burke T, Groenen MA, Crooijmans RP, Vignal A, Fillon V, Morisson M, Pitel F, Tixier-Boichard M, Ladjali-Mohammedi K, Hillel J, Maki-Tanila A, Cheng HH, Delany ME, Burnside J, Mizuno S (2000) First report on chicken genes and chromosomes. Cytogenet Cell Genet 90:169–218
Scholz B, Kultima K, Mattsson A, Axelsson J, Brunström B, Halldin K, Stigson M, Dencker L (2006) Sex-dependent gene expression in early brain development of chicken embryos. BMC Neurosci 7:12
Shetty S, Griffin DK, Graves JA (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res 7(4):289–295
Sokol SB, Kuwabara PE (2000) Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev 14(8):901–906
Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET (2007) A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell 13(1):127–139
Tsuda Y, Nishida-Umehara C, Ishijima J, Yamada K, Matsuda Y (2007) Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma 116(2):159–173
Vallender EJ, Lahn BT (2004) How mammalian sex chromosomes acquired their peculiar gene content. Bioessays 26(2):159–169
Ventura-Holman T, Seldin MF, Li W, Maher JF (1998) The murine fem1 gene family: homologs of the Caenorhabditis elegans sex-determination protein FEM-1. Genomics 54(2):221–230
Ventura-Holman T, Lu D, Si X, Izevbigie EB, Maher JF (2003) The FEM1C genes: conserved members of the Fem1 gene family in vertebrates. Gene 314:133–139
Acknowledgments
This work was supported by NIH grant DC000217 to A.P. Arnold and a Yamada Science Foundation grant to Y. Itoh. BAC sequences were determined by the Washington University Genome Center under the supervision of Dr. Wes Warren. We thank Klaus-Peter Koepfli and Robert Wayne for the helpful discussions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Comai
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Itoh, Y., Kampf, K. & Arnold, A.P. Disruption of FEM1C-W gene in zebra finch: evolutionary insights on avian ZW genes. Chromosoma 118, 323–334 (2009). https://doi.org/10.1007/s00412-008-0199-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-008-0199-8