Abstract
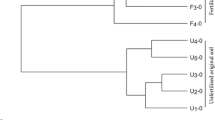
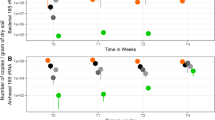
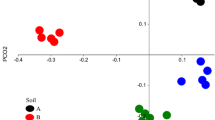
Soil is considered as one of the most biodiverse environments on Earth; yet, the taxonomy, occurrence, and role of its different microbial populations are largely unknown. Here, two sterilized soils (from England and Italy) were inoculated with a subsample of their initial microbial communities and/or those from the other soil to study their microbial community evolution. This approach compared two driving factors (original community and soil physico-chemical characteristics) for microbial community definition. After 2 months of incubation and based on metagenomic datasets, the two inoculated communities (from an English grassland and an Italian forest) possessed similar functional and taxonomical structures when inoculated in the same sterile soil. For example, the newly colonized Italian soil was dominated by Actinobacteria related organisms (>66 % of the detected community) with a functional distribution independent of the inoculated soil origin. In addition, some of the organisms that dominated the different inoculated communities after 2 months were similar for a given sterile soil whether they came from the English grassland or the Italian forest, and they had not been detected in the original microbial community from either soil. Thus, similar microorganisms with low representation from the two distinct communities emerged in each sterilized soil, thus increasing the microbial diversity recovered from the microbial community of the donor soil. So far, these observations support the idea that different temperate soil microbial communities have different evenness due to environmental physico-chemical variations, yet have similar community composition (richness), and thus develop similarly when colonizing the same habitat.






Similar content being viewed by others
References
Ashby MN, Rine J, Mongodin EF, Nelson KE, Dimster-Denk D (2007) Serial analysis of rRNA genes and the unexpected dominance of rare members of microbial communities. Appl Environ Microbiol 73:4532–4542
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM (2012) Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 48:489–499
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM, Ribosomal Database P (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Delmont TO, Malandain C, Prestat E, Larose C, Monier JM, Simonet P, Vogel TM (2011a) Metagenomic mining for microbiologists. ISME J 5:1837–1843
Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, Hirsch PR, Vogel TM (2011b) Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol 77:1315–1324
Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM, Pelletier E, Hirsch PR, Meyer F, Gilbert JA, Le Paslier D, Simonet P, Vogel TM (2012) Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J 6:1677–1687
Delmont TO, Simonet P, Vogel TM (2013) Mastering methodological pitfalls for surviving the metagenomic jungle. BioEssays 35:744–754
Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO, Roe BA, Davis JP, Schloss PD, Bailey VL, Krumholz LR (2008) Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol 74:5422–5428
Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395
Gans J, Wolinsky M, Dunbar J (2005) Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390
Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491
Hamp TJ, Jones WJ, Fodor AA (2009) Effects of experimental choices and analysis noise on surveys of the “rare biosphere”. Appl Environ Microbiol 75:3263–3270
Hong S, Bunge J, Leslin C, Jeon S, Epstein SS (2009) Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3:1365–1373
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma 9:386
Morales SE, Cosart TF, Johnson JV, Holben WE (2009) Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl Environ Microbiol 75:668–675
Niu B, Fu L, Sun S, Li W (2010) Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinforma 11:187
Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702
Parks DH, Beiko RG (2010) Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26:715–721
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Soergel DA, Dey N, Knight R, Brenner SE (2012) Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J 6:1440–1444
Torsvik V, Ovreas L, Thingstad TF (2002) Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064–1066
Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, Van Elsas JD, Bailey MJ, Nalin R, Philippot L (2010) TerraGenome: a consortium for the sequencing of a soil metagenome. Nature reviews Microbiology p 252
Acknowledgment
We would like to thank Penny Hirsch and Ian Clark for providing the soil from Rothamsted, as well as Denis Le Paslier and Eric Pelletier for their involvement in generating metagenomic datasets at the Genoscope. This research was supported by the French National Research Agency (Agence National de Recherche) project Metasoil (Projet ANR-08-GENM-025). TOD was funded by the Rhone-Alpes Région.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Box plots of variance based on the relative number of sequenced reads annotated into the specific subsystem for each of the nine experiments (see material and method section) in replicates after 8 weeks in microcosms and clustered by group. (GIF 3588 kb)
Fig. S2
Box plots of variance based on the relative number of sequenced reads annotated into the specific subsystem for each of the nine experiments (see material and method section) in replicates after 8 weeks in microcosms and clustered by group. (GIF 4296 kb)
Rights and permissions
About this article
Cite this article
Delmont, T.O., Francioli, D., Jacquesson, S. et al. Microbial community development and unseen diversity recovery in inoculated sterile soil. Biol Fertil Soils 50, 1069–1076 (2014). https://doi.org/10.1007/s00374-014-0925-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-014-0925-8