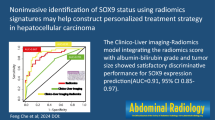
Abstract
Objectives
Immunoscore evaluates the density of CD3+ and CD8+ T cells in both the tumor core and invasive margin. Pretreatment prediction of immunoscore in hepatocellular cancer (HCC) is important for precision immunotherapy. We aimed to develop a radiomics model based on gadolinium-ethoxybenzyl-diethylenetriamine (Gd-EOB-DTPA)-enhanced MRI for pretreatment prediction of immunoscore (0–2 vs. 3–4) in HCC.
Materials and methods
The study included 207 (training cohort: n = 150; validation cohort: n = 57) HCC patients with hepatectomy who underwent preoperative Gd-EOB-DTPA-enhanced MRI. The volumes of interest enclosing hepatic lesions including intratumoral and peritumoral regions were manually delineated in the hepatobiliary phase of MRI images, from which 1044 quantitative features were extracted and analyzed. Extremely randomized tree method was used to select radiomics features for building radiomics model. Predicting performance in immunoscore was compared among three models: (1) using only intratumoral radiomics features (intratumoral radiomics model); (2) using combined intratumoral and peritumoral radiomics features (combined radiomics model); (3) using clinical data and selected combined radiomics features (combined radiomics-based clinical model).
Results
The combined radiomics model showed a better predicting performance in immunoscore than intratumoral radiomics model (AUC, 0.904 (95% CI 0.855–0.953) vs. 0.823 (95% CI 0.747–0.899)). The combined radiomics-based clinical model showed an improvement over the combined radiomics model in predicting immunoscore (AUC, 0·926 (95% CI 0·884–0·967) vs. 0·904 (95% CI 0·855–0·953)), although differences were not statistically significant. Results were confirmed in validation cohort and calibration curves showed good agreement.
Conclusion
The MRI-based combined radiomics nomogram is effective in predicting immunoscore in HCC and may help making treatment decisions.
Key Points
• Radiomics obtained from Gd-EOB-DTPA-enhanced MRI help predicting immunoscore in hepatocellular carcinoma.
• Combined intratumoral and peritumoral radiomics are superior to intratumoral radiomics only in predicting immunoscore.
• We developed a combined clinical and radiomicsnomogram to predict immunoscore in hepatocellular carcinoma.





Similar content being viewed by others
Abbreviations
- AFP:
-
Alpha-fetoprotein
- AST:
-
Aspartate transaminase
- CT:
-
Center of the tumor
- DAB:
-
Diaminobenzidine
- DCA:
-
Decision curve analysis
- Gd-EOB-DTPA:
-
Gadolinium-ethoxybenzyl-diethylenetriamine
- GGT:
-
γ-Glutamyl transpeptadase
- GLCM:
-
Gray level co-occurrence matrix
- GLRCM:
-
Gray level run-length matrix
- HBP:
-
Hepatobiliary phase
- HCC:
-
Hepatocellular carcinoma
- ICB:
-
Immune checkpoint blockade
- ICC:
-
Intra-class correlation coefficient
- IM:
-
Invasive margin
- NPV:
-
Negative predictive value
- PD-1:
-
Programmed death receptor 1
- PD-L1:
-
Programmed death-ligand 1
- PPV:
-
Positive predictive value
- TIL:
-
Tumor infiltrating lymphocytes
- TME:
-
Tumor microenvironment
- VOI:
-
Volumes of interest
References
Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S (2015) Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg 261:947–955
El-Khoueiry AB, Sangro B, Yau T et al (2017) Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389:2492–2502
Sangro B, Gomez-Martin C, de la Mata M et al (2013) A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol 59:81–88
Palucka AK, Coussens LM (2016) The basis of oncoimmunology. Cell 164:1233–1247
Kim YJ (2015) Subverting the adaptive immune resistance mechanism to improve clinical responses to immune checkpoint blockade therapy. Oncoimmunology 3:e954868
Taube JM (2014) Unleashing the immune system: PD-1 and PD-Ls in the pre-treatment tumor microenvironment and correlation with response to PD-1/PD-L1 blockade. Oncoimmunology 11:e963413
Zhou G, Sprengers D, Boor PPC et al (2017) Antibodies against immune checkpoint molecules restore functions of tumor-infiltrating T cells in hepatocellular carcinomas. Gastroenterology 153:1107–1119
Galon J, Mlecnik B, Bindea G et al (2014) Towards the introduction of the ‘immunoscore’ in the classification of malignant tumours. J Pathol 232:199–209
Galon J, Costes A, Sanchez-Cabo F et al (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313:1960–1964
Mahmoud SM, Paish EC, Powe DG et al (2011) Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 29:1949–1955
Brunner SM, Rubner C, Kesselring R et al (2015) Tumor-infiltrating, interleukin-33–producing effector-memory CD8+ T cells in resected hepatocellular carcinoma prolong patient survival. Hepatology 61:1957–1967
Jiang Y, Zhang Q, Hu Y et al (2018) ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg 267:504–513
Donnem T, Hald SM, Paulsen EE et al (2015) Stromal CD8+ T-cell density-a promising supplement to TNM staging in non-small cell lung cancer. Clin Cancer Res 21:2635–2643
Gabrielson A, Wu Y, Wang H et al (2016) Intratumoral CD3 and CD8 T-cell densities associated with relapse free survival in HCC. Cancer Immunol Res 4:419–430
Sun C, Xu J, Song J et al (2015) The predictive value of centre tumour CD8+ T cells in patients with hepatocellular carcinoma: comparison with immunoscore. Oncotarget 6:35602–35615
Yao Q, Bao X, Xue R et al (2017) Prognostic value of immunoscore to identify mortality outcomes in adults with HBV-related primary hepatocellular carcinoma. Medicine (Baltimore) 96(17):e67356
Garnelo M, Tan A, Her Z et al (2017) Interaction between tumor-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut 66:342–351
Shields AF, Jacobs P, Sznol M et al (2018) Immune modulation therapy and imaging: workshop report. J Nucl Med 59:410–417
Ku YJ, Kim HH, Cha JH et al (2016) Correlation between MRI and the level of tumor-infiltrating lymphocytes in patients with triple negative breast cancer. AJR Am J Roentgenol 207:1146–1151
Ku YJ, Kim HH, Cha JH et al (2018) Predicting the level of tumor-infiltrating lymphocytes in patients with triple negative breast cancer: usefulness of breast MRI computer-aided detection and diagnosis. J Magn Reson Imaging 47:760–766
Kumar V, Gu Y, Basu S et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248
Sun R, Limkin EJ, Dercle L et al (2017) Computational medical imaging (radiomics) and potential for immuno-oncology. Cancer Radiother 21:648–654
Savadjiev P, Chong J, Dohan A et al (2018) Demystification of AI-driven medical image interpretation: past, present and future. Eur Radiol. https://doi.org/10.1007/s00330-018-5674-x
Li Y, Liu X, Xu K et al (2018) MRI features can predict EGFR expression in lower grade gliomas: a voxel-based radiomic analysis. Eur Radiol 28:356–362
Esteva A, Kuprel B, Novoa RA et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118
Braman NM, Etesami M, Prasanna P et al (2017) Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 19:57
Rutman AM, Kuo MD (2009) Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 70:232–241
Wu M, Tan H, Gao F et al (2018) Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol. https://doi.org/10.1007/s00330-018-5787-2
Hamm B, Staks T, Mühler A et al (1995) Phase I clinical evaluation of Gd-EOB-DTPA as a hepatobiliary MR contrast agent: safety, pharmacokinetics, and MR imaging. Radiology 195:785–792
Choi JW, Lee JM, Kim SJ et al (2013) Hepatocellular carcinoma: imaging patterns on gadoxetic acid-enhanced MR images and their value as an imaging biomarker. Radiology 267:776–786
Zhang B, Tian J, Dong D et al (2017) Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res 23:4259–4269
Bakr S, Echegaray S, Shah R et al (2017) Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham) 4:041303
Huang YQ, Liang CH, He L et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157–2164
Li H, Zhu Y, Burnside ES et al (2016) MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 281:382–391
Galon J, Pagès F, Marincola FM et al (2012) Cancer classification using the immunoscore: a worldwide task force. J Transl Med 10:205
Leijenaar RT, Carvalho S, Velazquez ER et al (2013) Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 52:1391–1137
Geurts P, Ernst D, Wehenkel L (2006) Extremely randomized trees. Mach Learn 63:3–42
Marée R, Geurts P, Wehenkel L (2007) Random subwindows and extremely randomized trees for image classification in cell biology. BMC Cell Biol 8(Suppl 1):S2
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Bourgier C, Colinge J, Aillères N et al (2015) Radiomics: definition and clinical development. Cancer Radiother 19:532–537
Grossmann P, Stringfield O, El-Hachem N et al (2017) Defining the biological basis of radiomic phenotypes in lung cancer. Elife 6. https://doi.org/10.7554/eLife.23421
Fox MJ, Gibbs P, Pickles MD (2016) Minkowski functionals: an MRI texture analysis tool for determination of the aggressiveness of breast cancer. J Magn Reson Imaging 43:903–910
Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA (2013) Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 266:326–336
Segal E, Sirlin CB, Ooi C et al (2007) Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 25:675–680
Funding
This study has received funding by grants from the Guangzhou Science and Technology Program key projects (No. 201803010057) and the National Natural Science Foundation of China (No. 81771908, 81571750). This work was supported by Ministry of Science and Technology of China under Grant No. 2017YFA0205200, National Natural Science Foundation of China under Grant No. 81227901, 81527805, Chinese Academy of Sciences under Grant No. GJJSTD20170004 and QYZDJ-SSW-JSC005, Beijing Municipal Science & Technology Commission under Grant No. Z161100002616022, Z171100000117023, the Key International Cooperation Projects of the Chinese Academy of Sciences under Grant No. 173211KYSB20160053. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Ming Kuang.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
Two of the authors (Fei Liu, Bin Li) have significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 12520 kb)
Rights and permissions
About this article
Cite this article
Chen, S., Feng, S., Wei, J. et al. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol 29, 4177–4187 (2019). https://doi.org/10.1007/s00330-018-5986-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5986-x