Abstract
Key message
Tetraploid `Moncada´ mandarin, used as male and female in interploidy hybridizations, displays mainly tetrasomic inheritance for most LGs, with slight variations according to the direction of the crossing.
Abstract
Triploid-breeding programs in citrus are key tool to develop seedless cultivars. Obtaining triploid citrus hybrids may be achieved through different strategies, such as the exploitation of female unreduced gamete in crosses between diploid parents and diploid by tetraploid sexual hybridizations, in which tetraploid genotypes can be used as male or female parents. Genetic configuration of triploid populations from interploid crosses greatly depends on the chromosomic segregation mode of the tetraploid parent used. Here, we have analyzed the inheritance of the tetraploid ‘Moncada’ mandarin and compared the genetic structures of the resulting gametes when used as male and as female parent. The preferential chromosome pairing rate is calculated from the parental heterozygosity restitution (PHR) of codominant molecular markers, indicating the proportion between disomic and tetrasomic segregation. Tetraploid ‘Moncada’ both as female and male parent largely exhibited tetrasomic segregation. However, as female parent, one linkage group (LG8) showed intermediate segregation with tendency towards tetrasomic inheritance, while another linkage group (LG4) evidenced a clear intermediate segregation. On the other hand, when used as male parent two linkage groups (LG5 and LG6) showed values that fit an intermediate inheritance model with tetrasomic tendency. Significant doubled reduction (DR) rates were observed in five linkage groups as female parent, and in six linkage groups as male parent. The new knowledge generated here will serve to define crossing strategies in citrus improvement programs to efficiently obtain new varieties of interest in the global fresh consumption market.
Similar content being viewed by others
Introduction
Polyploids are plants with somatic cells that contain three or more complete sets of chromosomes (Ramsey and Schemske 1998). Ancient whole-genome duplications have been reported in most evolutionary lineages and may represent a crucial mode of speciation and eukaryotic genome evolution (Cai et al. 2019; Van de Peer et al. 2017). In fact, all the angiosperm genomes sequenced to date exhibit evidence of ancient polyploidization events (Cai et al. 2019; Soltis et al. 2014; Van de Peer et al. 2017) and polyploidy is one of the major forces of evolution for plant species, leading to their diversification and differentiation (Gallais 2003; Otto and Whitton 2000; Van de Peer et al. 2017).
Basically, polyploids differ from the diploid counterparts in their ecological, morphological, and physiological characteristics (Dewitte et al. 2009; Guerra et al. 2014; Ramsey 2007; Ruiz et al. 2016). Several mechanisms lead to polyploidy, such as somatic doubling or the production of unreduced gametes which is the main polyploidization mechanism reported in plants (Bretagnolle and Thompson 1995; De Storme and Geelen 2013; Ramsey and Schemske 1998, 2002).
Polyploidization offers many opportunities as a valuable tool in citrus-breeding programs (Aleza et al. 2016; Cuenca et al. 2015; Grosser and Gmitter 2011; Ollitrault et al. 2008). In Citrus and related genera, diploid genotypes are the most common, with a basic chromosome number x = 9 (Krug 1943). However, euploids and aneuploids have been induced or found occasionally, with triploids and tetraploids being the most common euploid variations (Lee 1988). Citrus triploid genotypes are generally seedless, a demanded characteristic for fresh fruit marketing (Aleza et al. 2012a, b, 2016). However, a few seedy triploid lime varieties have been described (Curk et al. 2016). Triploid genotypes in citrus are routinely obtained by sexual hybridization, through unreduced female gametes (Aleza et al. 2016; Cuenca et al. 2011, 2015), and interploid hybridizations between diploid and tetraploid genotypes (Aleza et al. 2012a, b; Grosser and Gmitter 2011; Starrantino and Recupero 1982).
There are two extreme models for diploid gametes produced by tetraploid plants, i.e., disomic in allotetraploids and tetrasomic in autotetraploids (Stebbins 1947; Stift et al. 2008; Sybenga 2012). The fusion of the genomes of two species gives rise to the allotetraploids, which present two sets of homologous chromosomes. During meiosis, each chromosome is paired with its homologous and forms only bivalents (Stebbins 1947; Sybenga 2012). This generates a 100% interspecific heterozygosity transmitted by each gamete, resulting in a disomic inheritance (Stift et al. 2008). In contrast, the four homologous chromosomes in the autotetraploids have the same opportunity to mate during meiosis, leading to multivalent formation and thus, tetrasomic inheritance (Aleza et al. 2016; Jackson and Jackson 1996; Sybenga 1996). For autotetraploids resulting from somatic chromosome doubling of diploid varieties, it theoretically leads to 66% restitution of the heterozygosity of the diploid that originates the tetraploid (Aleza et al. 2016; Sanford 1983). In fact, allo- and autotetraploids are the extremes of the range. In cases where parents are divergent, but have retained enough homology to prevent exclusive preferential pairing, inheritance patterns intermediate between di- and tetrasomic can be expected (Jeridi et al. 2012; Stebbins 1947; Stift et al. 2008; Sybenga 1996). Intermediate inheritance patterns have been revealed in citrus allotetraploid somatic hybrids (Kamiri et al. 2011, 2018) and for the tetraploid ‘Clemenules’ clementine (Aleza et al. 2016). Stift et al. (2008) developed a likelihood-based approach to evaluate whether disomic, intermediate, or tetrasomic inheritances best fitted the segregation of genetic markers and to estimate preferential pairing and double reduction (DR) rates. DRs can occur when tetravalent are formed and increase the homozygosity of diploid gametes (Aleza et al. 2016; Ronfort et al. 1998; Stift et al. 2008; Sybenga 1996). A simplified likelihood method was proposed by Aleza et al. (2016) for tetraploid resulting from somatic chromosome doubling.
Molecular marker analysis indicate that cultivated citrus resulted from complex interspecific admixtures of four ancestral taxa: C. reticulata (mandarin), C. maxima (pummelo), C. medica (citron), and C. micrantha (papeda) that arose during the domestication of citrus fruits (Curk et al. 2016; Froelicher et al. 2011; Garcia-Lor et al. 2013b; Nicolosi et al. 2000) and these results were confirmed by sequencing data (Wu et al. 2014, 2018; Xu et al. 2013). Commonly, the tetraploid parents used in interploid hybridizations for triploid breeding result from somatic chromosome doubling occurring spontaneously in nucellar cells or induced by treatment using antimitotic agents such as colchicine and oryzaline (Aleza et al. 2009, 2011). In relation with the phylogenetic origin of the parental diploid such somatic tetraploids can be autotetraploid for monospecific varieties, allotetraploids when parental diploid resulted from direct interspecific hybridization or segmental allotetraploid when parental diploid had a more complex admixture genome. These complex genomes may, therefore, impact the observed segregations in breeding programs.
Here, we analyze the segregation pattern of the tetraploid ‘Moncada’ mandarin used both as male and as female parent in interploid crosses by genotyping triploid progenies with Simple Sequence Repeat (SSR) and Single-Nucleotide Polymorphism (SNP) molecular markers.
Diploid ‘Moncada’ mandarin was obtained from after 1980 in a breeding program held at Instituto Valenciano de Investigaciones Agrarias (IVIA) from a handmade pollination between ‘Oroval’ clementine (Citrus clementina Hort. Ex Tan.) and ‘Kara’ mandarin (C. unshiu (Mak) Marc. × C. nobilis Lour.) (Bermejo et al. 2011). Later, tetraploid ‘Moncada’ mandarin was obtained by colchicine treatment of shoot tips grafted in vitro (Aleza et al. 2009). This mandarin hybrid is characterized by its excellent fruit quality, very easy to peel, very late maturity period and also is a non-apomictic genotype what makes a very interesting parent in citrus-breeding programs based on sexual hybridizations aimed to recover large populations of triploid hybrids. The breeding implications of the use of the tetraploid ‘Moncada’ mandarin as male or female parent in the recovery of large populations of triploid hybrids are further discussed.
Materials and methods
Plant material
Triploid hybrid progenies were obtained from 4 × × 2× and 2× × 4× sexual hybridizations using tetraploid ‘Moncada’ mandarin as female and male parent, respectively. Tetraploid ‘Moncada’ mandarin was obtained directly from shoot tip grafting combined with colchicine treatment (Aleza et al. 2009). In 4 × × 2× sexual hybridization, 72 triploid hybrids were recovered using diploid ‘Anana’ mandarin (C. reticulata) as male parent (from here on referred as MA hybridization), whereas in the 2 × × 4× sexual hybridization, 88 triploid hybrids were obtained with the non-apomictic diploid ‘Clemenules’ clementine female parent (from here on referred as CM hybridization). Ploidy-level analysis by flow cytometry and triploid hybrid recovery was performed following the methodology described by Aleza et al. (2012a, b).
Genotyping of the triploid progenies
To study the genetic structure of the diploid gametes produced by the tetraploid ‘Moncada’ mandarin, progenies along with the parents were genotyped using SSR and SNP markers distributed homogeneously in the nine linkage groups (LGs) of the clementine reference genetic map (Ollitrault et al. 2012a). These markers were heterozygous for ‘Moncada’ mandarin and displayed polymorphism between ‘Moncada’ mandarin and ‘Clemenules’ or ‘Anana’ mandarins. Since ‘Moncada’ is a direct hybrid between clementine and ‘Kara’ mandarin, it was difficult to find heterozygous markers for ‘Moncada’ mandarin with polymorphism with clementine. Finally, 24 SSRs and 19 SNPs markers previously developed were analyzed for both populations. In addition, 11 new SNP markers were developed (Table 1) from a Genotyping-by-Sequencing (GBS) diversity analysis (unpublished data). Detailed information about SSR and SNP markers used in this study is given in Table 2. Given the genetic proximity between the tetraploid ‘Moncada’ and clementines, the exact same set of molecular markers could not be used in both families (CM and MA). Even so, 13 molecular markers were used in common for both families, distributed in eight out of the nine LGs.
PCR amplifications using SSR markers were performed using a thermocycler rep gradient S (Eppendorf®) in 15 μL containing 0.5 μl 1U/μl of Taq DNA polymerase (Fermentas ®), 3 μL citrus DNA, 1.5 μl of 2 mM welled (Sigma ®) dye-labeled forward primer, 1.5 μl of 2 mM non-dye-labeled reverse primer, 0.2 mM of each dNTP, 1.5 μl 10× PCR buffer, and 0.45 μl 50 mM MgCl2. The PCR protocol was as follows: denaturation at 94 °C for 5 min followed by 40 cycles of 30 s at 94 °C, 30 s at 50 or 55 °C, and 30 s at 72 °C; and a final elongation step of 8 min at 72 °C. Capillary electrophoresis was carried out using a Genetic Analysis System 8000 (Beckman Coulter Inc.). The PCR products were initially denatured at 90 °C for 2 min, loaded at 2 kV for 30 s, and separated at 6 kV for 35 min. Alleles were sized based on a DNA size standard (400 bp). GenomeLab™ v.10.0 (Beckman Coulter Inc.) genetic analysis software was used for data collection.
SNP markers were genotyped using KASPar™ technology by LGC Genomics (Hoddesdon, UK). The KASPar™ genotyping system is a competitive, allele-specific dual Förster resonance energy transfer (FRET)-based assay for SNP genotyping. Primers were directly designed by LGC Genomics based on the SNP locus flanking sequence. Detailed explanation of the specific conditions and reagents used in KASPar™ technique can be found in Cuppen (2007). The allelic dose estimation in the heterozygous triploid hybrids was performed as described by Cuenca et al. (2013).
Data analysis
Inferring the diploid gamete genetic configuration
In interploid crosses leading to triploid progenies, diploid gametes are transmitted from the tetraploid parent (Aleza et al. 2012a, b). For loci with completely different parental allelic configurations (A1A2 × A3A4), the genotype of the 2× gamete can be read directly from the configuration of triallelic triploid hybrids. When the female and male parents share one allele (A1A2 × A2A2 or A1A2 × A2A3), we inferred the structure of the 2× gamete forming biallelic triploid hybrids from the allelic dose, as described by Cuenca et al. (2011, 2013). We confirmed that all triploid hybrids were formed through the fusion of a diploid gamete from the tetraploid parent and a haploid gamete from the diploid parent by either observing triallelic configuration in the hybrids for at least one marker or from dosage estimation.
Parental heterozygosity restitution (PHR)
The PHR was calculated for each locus as the percentage of triploid individuals with the heterozygous allelic configuration inherited from tetraploid ‘Moncada’ mandarin transmitted through diploid gametes. Similarly, PHR was calculated for each individual as the percentage of loci with the same heterozygous allelic configuration as tetraploid ‘Moncada’ mandarin.
Estimation of preferential association frequency and maximum double reduction rate
For citrus, Stift et al. (2008) proposed a segregation model for allotetraploids, which was simplified by Aleza et al. (2016) for tetraploid resulting from somatic chromosome doubling. It is considered that in such tetraploid, for centromeric loci, the expected frequencies of each type of gamete depend only on the ‘tetrasomic’ parameter (τ), corresponding to the proportion of gametes formed by random associations of meiotic chromosomes (i.e., random bivalent or tetravalent pairing). The estimation of τ was performed using a maximum likelihood approach from the analysis of the marker closest to the centromere for each LG, as proposed by Aleza et al. (2016). This value ranges from 0 for completely disomic to 1 for complete tetrasomic inheritance. Confidence intervals (CIs) were estimated following a similar approach to the LOD drop-off method (Lander and Botstein 1989), by finding the values at either side of the estimated τ that corresponded to a tenfold decrease in probability. Then, preferential pairing (PP) was calculated as 1 − τ.
The double reduction rate (DR) and its confidence interval (CI) for each LG were estimated as proposed by Aleza et al. (2016). Briefly, DR is estimated from τ values for each LG for the markers furthest from the centromere applying a maximum likelihood approach, and the CI corresponds to the values on each side with a tenfold decrease in the probability.
Population diversity organization
Genetic differences between individuals were estimated using the DARwin6 software (Perrier and Jacquemound-Collet 2018) and analyzed with a neighbor-joining analysis using the simple matching dissimilarity index):
where \(d_{i - j}\) is the dissimilarity between units i and j, L is the number of loci, \(m_{l}\) is the number of matching alleles for locus l, and \(\pi\) is the ploidy. From the dissimilarity matrix obtained, a weighted neighbor-joining tree (Saitou and Nei 1987) was computed.
The potential distortion in allelic segregation was analyzed using Chi-square test (χ2) with the Bonferroni correction for multiple testing applied (Bonferroni 1936; Goeman and Solari 2014; Holm 1979).
For group differentiation between the analyzed triploid hybrids of each progeny, the G/N relation was used, where G is the number of groups differentiated by the molecular markers used within each LG, and N is the total number of genotypes. The groups were obtained with the DARwin6 software (Perrier and Jacquemound-Collet 2018).
Results and discussion
Triploid genotyping
The genotyping of the triploid progenies was performed with 36 markers for MA and 31 for CM hybridizations, which allowed the unequivocal allelic differentiation between both parents and the determination of the origin of the diploid gametes that gave rise to each triploid hybrid.
Triallelic configurations with two alleles arising from tetraploid ‘Moncada’ were observed for all hybrids from MA for at least one SSR marker, directly confirming that the 2× gametes came from the tetraploid ‘Moncada’ progenitor. However, for CM hybrids, all molecular markers showed biallelic configurations, and the allele dosages were estimated as proposed by Cuenca et al. (2015). Finally, all triploid hybrids in both families were confirmed to arise from the fusion of a diploid gamete from tetraploid ‘Moncada’ and a haploid gamete from the diploid genitor (Fig. 1). Once the origin of the 2× gametes was confirmed, their genetic configurations were inferred for all marker-gamete combinations (Supplementary Table 1). An example for assessing genetic configuration from the direct observation of triallelic hybrids and the dosage estimation the peak ratio from a triallelic hybrid for the CI01F04a SSR marker is given in Fig. 1. In this case, tetraploid ‘Moncada’ shows 186/210 alelles (Fig. 1a) and ‘Anana’ 199/201 alleles (Fig. 1b). Hybrid ‘MA14’ shows 186/199/210 allele configuration (Fig. 1c), thus allows directly inferring 186/210 configuration for the 2× gamete from tetraploid ‘Moncada’ (heterozygosity restitution). In contrast, the hybrid ‘MA50’ for the same marker shows 199/210 allelic configuration (Fig. 1d), and therefore, the allelic dose estimation was done considering the relationship between the alleles 199/210 of the triallelic triploid hybrid as a baseline. It was concluded a 199/210:210 genotype for ‘MA50′ and consequently 210/210 genotypes for the 2× gamete from tetraploid ‘Moncada’ (no heterozygosity restitution).
The potential distortion in allelic segregation for the two types of homozygous gametes was analyzed using Chi-square test (χ2) with the Bonferroni correction for multiple testing applied. Only the marker MEST256 in LG3 for the MA population (Table 3) and the markers CHS-M183, MEST123 and FLS-M400 in LG3, LG6, and LG7, respectively, for the CM population presented distortion in allele segregations (Table 4).
Other citrus studies showed segregation distortions. Bernet et al. (2010) analyzed reciprocal crosses between ´Fortune´ mandarin and ´Chandler´ pummelo, obtaining progenies with allelic frequencies distorted in both populations. In the same way, Ollitrault et al. (2012a) observed segregation distortions in male and female gametes of ´Clemenules’ clementine. In both studies, distortions were higher for the male gametes and the authors suggested that general factors such as mechanisms of gamete abortion, pollen competition, or gametophytic incompatibility could be related with them (Bernet et al. 2010; Ollitrault et al. 2012a).
Genetic structure of diploid gamete populations arising from tetraploid ‘Moncada’ mandarin as female and male parent
Variability of PHR
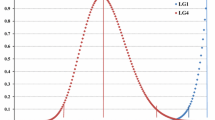
The PHR obtained from tetraploid ‘Moncada’ as male and female parent was calculated at gamete and marker level. At the gamete level, PHR presented a unimodal distribution when tetraploid ‘Moncada’ was used as female parent (Fig. 2), with a PHR average of 0.654 ± 0.093. The unimodal distribution observed in tetraploid ‘Moncada’ as female parent was similarly observed for tetraploid ‘Clemenules’ clementine analyzed by Aleza et al. (2016). In contrast, a more heterogeneous distribution was observed when used as male parent, displaying 14 diploid gametes (‘CM19’, ‘CM21’, ‘CM25’, ‘CM48’, ‘CM54’, ‘CM55’, ‘CM60’, ‘CM73’, ‘CM74’, ‘CM75’, ‘CM78’, ‘CM83’, ‘CM85’, and ‘CM86’) with very low PHR values, ranging from 0.10 to 0.40. Therefore, the average of PHR was a little bit lower (0.599 ± 0.085) (Fig. 2). At marker level, both populations displayed a unimodal distribution of PHR, although the diploid male gamete population showed lower PHR values, probably originated by the diploid male gametes with low PHR values (Fig. 3).
MA produced 2× gametes with PHR values ranging from 0.528 for the CIC5842-02 SNP locus in LG5 to 0.833 for the CHI-M598 SNP locus in LG4 (Table 3). For the remaining LGs, PHR values remain mostly constant along the chromosome. On the other hand, CM produced 2× gametes with PHR values ranging from 0.432 for the TAA41 SSR locus in LG2 to 0.761 for the C6_1584763 SNP locus in LG6 (Table 4).
Comparing tetraploid ‘Moncada’ as female and male parent, the largest differences are found in LG 4 and 8. As female parent, PHR values were 0.794 ± 0.31 for LG4 and 0.74 ± 0.06 for LG 8; as male parent, PHR values were 0.614 ± 0.091 and 0.614 ± 0.041 for LG4 and 8, respectively (Table 5).
Genotypic variability
The genetic structure of these two populations was calculated by a neighbor-joining analysis (Fig. 4), allowing the differentiation of hybrid groups within each family and determine their genetic distance. The molecular markers used in this work made possible the differentiation of all triploid hybrids within each progeny (G/N = 1) (Table 5). The average genetic distance between gametes was slightly higher for CM (0.308 ± 0.0029) than for MA (0.278 ± 0.0027). In addition, the genetic structure of the MA population gametes is more homogeneous and compact than that obtained for the CM population. Comparing the genetic distances of both population gametes in relation to the tetraploid ‘Moncada’, CM displayed a genetic distance of 0.200 ± 0.093, whereas for MA, this distance was 0.173 ± 0.054. The results found for tetraploid ‘Moncada’, as male and female parent are consistent with those described by Aleza et al. (2016), which found a genetic distance value to tetraploid ‘Clemenules’ clementine of 0.176 ± 0.012 for the population of triploid hybrids obtained with this genotype as female parent. Nevertheless, in the CM gamete population, a group with higher genetic distance to the tetraploid ‘Moncada’ (0.362 ± 0.043) was observed (Fig. 4). This subpopulation is constituted by the same 14 diploid gametes described above with very low PHR. The genetic analysis performed in these hybrids reveals the same allele homozygosity configuration in nine (CIBE5720, C2_23768463, TAA41, CHSM183, C4P5278891, C4P25377913, Ci03D12a, Ci03B07, and C8P19129409) over the 31 molecular markers used, and also with two other SSR markers (MEST123 and Ci07D05) with the same homozygosity configuration except for only one diploid gamete. These molecular markers are located in all LGs, with the exception of LG9, and in the LG2 and LG6, three over the four markers analyzed in each LG, displayed the same allelic configuration in homozygosity.
Dendrograms corresponding to the genetic analysis performed with SSR and SNP markers obtained by calculating the Simple Matching Dissimilarity Index and construction of the tree by weighted neighbor-joining of two populations of triploid hybrids regenerated from crosses a tetraploid ‘Moncada’ × ‘Anana’ and b ‘Clemenules’ × tetraploid ‘Moncada’. The red circle highlights the position of tetraploid ‘Moncada’. The green circle highlights the group of hybrids furthest from tetraploid ‘Moncada’
Preferential pairing (PP) and maximum double reduction (DR)
The genome of many cultivated citrus is composed of mosaics of the ancestral species (Curk et al. 2014, 2015; Wu et al. 2014, 2018). The works carried out on citrus phylogeny (Oueslati et al. 2017; Wu et al. 2014, 2018) have shown that the genomes of the progenitors that gave rise to ‘Moncada’ mandarin (‘Oroval’ clementine (C. deliciosa × C. sinensis) and ‘Kara’ mandarin (C. unshiu × C. nobilis) are constituted by an interspecific mandarin/pummelo mosaic structure; therefore, ‘Moncada’ mandarin also has an interspecific structure in its chromosomes.
τ and PP were calculated for each LG from the segregation data of the markers closest to the centromere using the probability models (Aleza et al. 2016). These markers were located between 1.0 and 24.1 cM from the centromere. For tetraploid ‘Moncada’ as female parent (Table 6), complete tetrasomic inheritance was the best model for seven out of the nine LGs (LG1, LG2, LG3, LG5, LG6, LG7, and LG9). For LG8, an intermediate inheritance with tendency towards a tetrasomic inheritance (PP = 0.375) was estimated, while the LG4 evidenced a clear intermediate inheritance (PP = 0.5). For tetraploid ‘Moncada’ as male parent (Table 7), most of the chromosomes fit the tetrasomic inheritance model with the markers used, with PP = 0 for LG1, LG2, LG3, LG4, LG7, LG8, and LG9, while LG5 and LG6 showed values that fit an intermediate inheritance model with tetrasomic tendency (PP = 0.215 and 0.115, respectively).
Likewise, clementines also present an interspecific mandarin/pummelo structure (Wu et al. 2018) Aleza et al. (2016) studied the segregation model in tetraploid ‘Clemenules’ clementine as female parent, obtaining very similar results, as we report for the tetraploid ‘Moncada’ mandarin, generally fitting the tetrasomic inheritance model except for LG4, which fitted the intermediate inheritance model. However, they also reported that the LG6 and LG8 showed values that fit the intermediate inheritance model, with high tetrasomic tendency. Comparatively, we found that for ‘Moncada’ mandarin as female parent, the LG6 shows tetrasomic segregation, while results for the LG8 agree with the after as was reported for the tetraploid ‘Clemenules’ clementine, but with higher PP value. Subsequently, Rouiss et al. (2018) analyzed the segregation model of the tetraploid ‘Mexican’ lime (C. aurantiifolia), which originated from an interspecific hybridization between C. micrantha (papeda) and C. medica (Citron) (Curk et al. 2016; Nicolosi et al. 2000; Wu et al. 2018). The results showed that tetraploid ‘Mexican’ lime has intermediary inheritance with a preferential disomic trend. In addition, Kamiri et al. (2018) assessed the meiotic behavior of an intergeneric tetraploid somatic hybrid resulting from symmetric protoplast fusion of diploid C. reticulata and diploid Poncirus trifoliata, and observed an intermediate inheritance with a preferential disomic trend. On the other hand, the genotyping of the triploid progeny derived from a cross between diploid pummelo (C. maxima) and an allotetraploid intergeneric somatic hybrid between C. reticulata and C. limon showed a tetrasomic and intermediate inheritance for this citrus interspecific somatic hybrid (Kamiri et al. 2011). Altogether, these studies reveal that the preferential pairing of tetraploid citrus genotypes greatly varies in relation to their constitutive genomes. The differentiation between C. medica and C. micrantha as well as the one between C. reticulata and P. trifoliata seems to have a much more impact in preferential pairing than the one between C. maxima and C. reticulata. Tetraploid ´Moncada´ differs slightly in the segregation model when used as female or male parent. These sex-specific differences were also observed for salmon fish (Allendorf and Danzmann 1997). Disomic segregation was observed in females, while segregation in males was best explained by a mixture of disomic and tetrasomic inheritance.
The tetraploid ´Moncada´ as female parent showed significant values of DR in LG2, LG3, LG4, LG7, and LG9. For all LGs, the confidence intervals (CI) for DR values include the value of 1/6, considered as the maximum value of DR for tetrasomic segregation and one crossover event occurring between the marker and the corresponding centromere (Haynes and Douches 1993; Mather 1936; Bourke et al. 2015), although LGs 2 and 3 displayed a higher estimation of DR. When tetraploid ‘Moncada’ was used as male parent, significant values of DR were obtained for LG2, LG3, LG4, LG5, LG6, and LG7. For LG3, LG4, LG6, and LG7, the confidence intervals (CI) for DR values include the maximum value of DR under the hypothesis described above. In addition, LG2 and LG5 showed higher DR values. Tetraploid ‘Moncada´ shows the same trend as female and male parent in DR values for LG1, LG2, LG3, LG4, LG7, and LG8. The frequency of DR considers maximum values of 0 for random chromosome segregation hypothesis, 1/7 with pure random chromatid segregation hypothesis, and 1/6 with complete equational segregation (Mather 1935; Muller 1914). Estimated values over 1/6 should be due to the segregation distortion observed for the corresponding markers. Indeed, our model analysis is based on Mendelian segregation hypothesis, while negative sporophytic selection for dominant gene may induce a diminution of heterozygous frequencies (for the gene and linked markers) and results in overestimation of DR. Different works have been performed with the objective to estimate the DR frequency and these values have been ranged from 0 to almost 0.30 (Fisher 1947, 1950; Haynes and Douches 1993; Tai 1982a, b; Welch 1960; Wu et al. 2001). The values of DR rate can differ between loci according the tetrasomic inheritance model. This variability depends on both the chromosome in which the marker is located and the position of the marker within the chromosome. There are chromosomes with a greater tendency to form multivalent that would originate higher values of DR (Butruille and Boiteux 2000). In addition, DR could be better estimated using larger populations (Butruille and Boiteux 2000) and it is more probable to occur in markers located in telomeric rather than in centromeric regions, in which the probability of recombination events is close to zero (Aleza et al. 2015; Butruille and Boiteux 2000; Welch 1960). In addition, Butruille and Boiteux (2000) indicated that DR causes a decrease of the equilibrium frequencies of deleterious alleles, and it has much more influence on genes subjected to gametophytic selection than on genes solely under sporophytic selection. With gametophytic selection, low frequencies of DR are enough to reduce equilibrium frequencies several folds.
Implications for citrus-breeding programs
Two strategies are routinely exploited for obtaining citrus triploids, i.e., interploid hybridizations between 2× and 4× parents (Aleza et al. 2012a, b; Starrantino and Recupero 1982) and through female 2n gametes (Aleza et al. 2010; Cuenca et al. 2011, 2015). In interploid hybridizations, the tetraploid parent results usually from somatic chromosome doubling arising spontaneously in nucellar cells or induced by colchicine treatment. The study of the origin of the diploid gametes, which greatly influences the structure of the resulting triploid hybrid populations, is of great interest to select the most appropriate strategies to obtain new hybrids with desired characteristics. Cuenca et al. (2015) demonstrated that SDR mechanism gives rise to the 2n megagametophytes in diploid ‘Moncada’ mandarin. The use of this strategy produces hybrid progenies with large genetic variation, due to the relatively low transmission of the parental heterozygosity to the offspring (about 40% on average), thus resulting in high number of new allelic multilocus combinations. In this paper, we have analyzed the chromosome segregation in the tetraploid ‘Moncada’ mandarin, which showed predominantly tetrasomic segregation, when used both as female and male parent, with an average PHR of 65% when used as female and 60% as male parent. Moreover, PHR is relatively constant along the chromosomes. Therefore, if we compared with SDR-2n female gametes, interploid hybridizations with tetraploid ‘Moncada’ mandarin as tetraploid parent are potentially a more efficient strategy for the development of new varieties that are genotypically more similar to the ‘Moncada’ mandarin.
Furthermore, depending on the LG in which a gene controlling an eventual trait of interest is located, the genetic regulation of the trait and the direction of the crossing, different segregation in the offspring can be obtained. For example, the PHR in LG8 is higher when tetraploid Moncada is used as female than as male parent, and therefore, the progeny will show higher heterogeneity in this LG when using tetraploid Moncada as male parent. Considering a trait of interest controlled by a single dominant allele at a locus in LG8, the probability to obtain triploid hybrids that inherit the trait of interest is higher using tetraploid ‘Moncada’ as female parent.
Tetraploid ´Moncada´ mandarin displayed significant values of DR as male and female parent. DR results in a decrease of PHR and thus an increase of inbreeding (Haynes and Douches 1993). The production of higher levels of homozygosity could be useful in triploid mandarin breeding for the potential cleaning effect that DR can have by revealing deleterious alleles to selection (Butruille and Boiteux 2000; Bourke et al. 2015). DR also could increase the accumulation of rare but favorable allelic configurations through selection with molecular markers (Bourke et al. 2015).
The knowledge of the difference in segregations according to the crossing strategy (2n gametes or interploid hybridization) to obtain hybrid triploid progenies with the ‘Moncada’ mandarin opens a range of possibilities for designing efficient breeding programs aimed to obtain innovative products to fulfill the market demands.
Conclusions
The analysis of codominant marker segregation over the nine citrus chromosomes allowed to unravel the segregation pattern of the tetraploid ‘Moncada’. Using both as female and male parent, it displayed tetrasomic inheritance for most LGs, with slight variations according to the direction of the crossing. As female parent, LG8 showed intermediate inheritance with tendency towards tetrasomic inheritance, and LG4 evidenced clear intermediate inheritance. As male parent, LG5 and LG6 showed values that fit an intermediate inheritance model with tetrasomic tendency. Significant DR rates were found in LG2, LG3, LG4, LG7, and LG9 when using tetraploid Moncada as female parent and in LG2, LG3, LG4, LG5, LG6, and LG7 as male parent. Likewise, differences in PHR were found between tetraploid ‘Moncada’ as female parent and male parent, with higher values in LG 4 and LG 8 as female parent. The new knowledge generated here will serve to define crossing strategies in citrus improvement programs to efficiently obtain new varieties of interest in the global fresh consumption market.
References
Ahmad R, Struss D, Southwick SM (2003) Development and characterization of microsatellite markers in citrus. J Am Soc Hortic Sci 128:584–590. https://doi.org/10.21273/jashs.128.4.0584
Aleza P, Juárez J, Ollitrault P, Navarro L (2009) Production of tetraploid plants of non apomictic citrus genotypes. Plant Cell Rep 28:1837–1846. https://doi.org/10.1007/s00299-009-0783-2
Aleza P, Juárez J, Cuenca J, Ollitrault P, Navarro L (2010) Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2× × 2× sexual hybridisation and its application to extensive breeding programs. Plant Cell Rep 29:1023–1034. https://doi.org/10.1007/s00299-010-0888-7
Aleza P, Froelicher Y, Schwarz S, Agustí M, Hernández M, Juárez J et al (2011) Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Ann Bot 108:37–50. https://doi.org/10.1093/aob/mcr099
Aleza P, Juárez J, Cuenca J, Ollitrault P, Navarro L (2012a) Extensive citrus triploid hybrid production by 2× × 4× sexual hybridizations and parent-effect on the length of the juvenile phase. Plant Cell Rep 31:1723–1735. https://doi.org/10.1007/s00299-012-1286-0
Aleza P, Juárez J, Hernández M, Ollitrault P, Navarro L (2012b) Implementation of extensive citrus triploid breeding programs based on 4× × 2× sexual hybridisations. Tree Genet. Genomes 8:1293–1306. https://doi.org/10.1007/s11295-012-0515-6
Aleza P, Cuenca J, Hernández M, Juárez J, Navarro L, Ollitrault P (2015) Genetic mapping of centromeres in the nine Citrus clementina chromosomes using half-tetrad analysis and recombination patterns in unreduced and haploid gametes. BMC Plant Biol 15:80. https://doi.org/10.1186/s12870-015-0464-y
Aleza P, Cuenca J, Juárez J, Navarro L, Ollitrault P (2016) Inheritance in doubled-diploid clementine and comparative study with SDR unreduced gametes of diploid clementine. Plant Cell Rep 35:1573–1586. https://doi.org/10.1007/s00299-016-1972-4
Allendorf FW, Danzmann RG (1997) Secondary tetrasomic segregation of MDH-B and preferential pairing of homeologues in rainbow trout. Genetics 145:1083–1092
Bermejo A, Pardo J, Cano A (2011) Influence of gamma irradiation on seedless citrus production: pollen germination and fruit quality. Food Nutr. Sci. 02:169–180. https://doi.org/10.4236/fns.2011.23024
Bernet GP, Fernandez-Ribacoba J, Carbonell EA, Asins MJ (2010) Comparative genome-wide segregation analysis and map construction using a reciprocal cross design to facilitate citrus germplasm utilization. Mol Breed 25:659–673. https://doi.org/10.1007/s11032-009-9363-y
Bonferroni C (1936) Teoria statistica delle classi e calcolo delle probabilità. Pubbl. del R Ist. Super. di Sci. Econ. e Commer. di Firenze 8:3–62
Bourke PM, Voorrips RE, Visser RGF, Maliepaard C (2015) The double-reduction landscape in tetraploid potato as revealed by a high-density linkage map. Genetics 201(3):853–863. https://doi.org/10.1534/genetics.115.181008
Bretagnolle F, Thompson JD (1995) Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol 129:1–22. https://doi.org/10.1111/j.1469-8137.1995.tb03005.x
Butruille DV, Boiteux LS (2000) Selection-mutation balance in polysomic tetraploids: impact of double reduction and gametophytic selection on the frequency and subchromosomal localization of deleterious mutations. Proc Natl Acad Sci USA 97:6608–6613. https://doi.org/10.1073/pnas.100101097
Cai L, Xi Z, Amorim AM, Sugumaran M, Rest JS, Liu L et al (2019) Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytol 221:565–576. https://doi.org/10.1111/nph.15357
Cuenca J, Froelicher Y, Aleza P, Juárez J, Navarro L, Ollitrault P (2011) Multilocus half-tetrad analysis and centromere mapping in citrus: evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv “Fortune”. Heredity 107:462–470. https://doi.org/10.1038/hdy.2011.33
Cuenca J, Aleza P, Navarro L, Ollitrault P (2013) Assignment of SNP allelic configuration in polyploids using competitive allele-specific PCR: application to citrus triploid progeny. Ann Bot 111:731–742. https://doi.org/10.1093/aob/mct032
Cuenca J, Aleza P, Juárez J, García-Lor A, Froelicher Y, Navarro L et al (2015) Maximum-likelihood method identifies meiotic restitution mechanism from heterozygosity transmission of centromeric loci: application in citrus. Sci Rep 5:9897. https://doi.org/10.1038/srep09897
Cuppen E (2007) Genotyping by allele-specific amplification (KASPar). Cold Spring Harb. Protoc. pdb.prot4841. https://doi.org/10.1101/pdb.prot4841
Curk F, Ancillo G, Garcia-Lor A, Luro F, Perrier X, Jacquemoud-Collet J-P et al (2014) Next generation haplotyping to decipher nuclear genomic interspecific admixture in Citrus species: analysis of chromosome 2. BMC Genet 15:152. https://doi.org/10.1186/s12863-014-0152-1
Curk F, Ancillo G, Ollitrault F, Perrier X, Jacquemoud-Collet J-P, Garcia-Lor A et al (2015) Nuclear species-diagnostic SNP markers mined from 454 amplicon sequencing reveal admixture genomic structure of modern citrus varieties. PLoS One 10:e0125628. https://doi.org/10.1371/journal.pone.0125628
Curk F, Ollitrault F, Garcia-Lor A, Luro F, Navarro L, Ollitrault P (2016) Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann Bot 117:565–583. https://doi.org/10.1093/aob/mcw005
De Storme N, Geelen D (2013) Sexual polyploidization in plants—cytological mechanisms and molecular regulation. New Phytol 198:670–684. https://doi.org/10.1111/nph.12184
Dewitte A, Eeckhaut T, Van Huylenbroeck J, Van Bockstaele E (2009) Occurrence of viable unreduced pollen in a Begonia collection. Euphytica 168:81–94. https://doi.org/10.1007/s10681-009-9891-x
Fisher RA (1947) The theory of linkage in polysomic inheritance. Philos Trans R Soc Lond B Biol Sci 233:55–87. https://doi.org/10.1098/rstb.1947.0006
Fisher RA (1950) The Theory of Inbreeding. J. R. Stat. Soc. Ser. A 113:249. https://doi.org/10.2307/2981045
Froelicher Y, Dambier D, Bassene JB, Costantino G, Lotfy S, Didout C et al (2008) Characterization of microsatellite markers in mandarin orange (Citrus reticulata Blanco). Mol Ecol Resour 8:119–122. https://doi.org/10.1111/j.1471-8286.2007.01893
Froelicher Y, Mouhaya W, Bassene J-B, Costantino G, Kamiri M, Luro F et al (2011) New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny. Tree Genet Genomes 7:49–61. https://doi.org/10.1007/s11295-010-0314-x
Gallais A (2003) Quantitative genetics and breeding methods in autopolyploids plants, 2nd edn. INRA, France
Garcia-Lor A, Ancillo G, Navarro L, Ollitrault P (2013a) Citrus (Rutaceae) SNP markers based on competitive allele-specific PCR; transferability across the aurantioideae subfamily. Appl Plant Sci 1:1200406. https://doi.org/10.3732/apps.1200406
Garcia-Lor A, Curk F, Snoussi-Trifa H, Morillon R, Ancillo G, Luro F et al (2013b) A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Ann Bot 111:1–19. https://doi.org/10.1093/aob/mcs227
García-Lor A, Luro F, Navarro L, Ollitrault P (2012) Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Mol Genet Genomics 287:77–94. https://doi.org/10.1007/s00438-011-0658-4
Goeman JJ, Solari A (2014) Multiple hypothesis testing in genomics. Stat Med 33:1946–1978. https://doi.org/10.1002/sim.6082
Grosser JW, Gmitter FG (2011) Protoplast fusion for production of tetraploids and triploids: applications for scion and rootstock breeding in citrus. Plant Cell, Tissue Organ Cult 104:343–357. https://doi.org/10.1007/s11240-010-9823-4
Guerra D, Wittmann MTS, Schwarz SF, de Souza PVD, Gonzatto MP, Weiler RL (2014) Comparison between diploid and tetraploid citrus rootstocks: morphological characterization and growth evaluation. Bragantia 73:1–7. https://doi.org/10.1590/brag.2014.007
Haynes KG, Douches DS (1993) Estimation of the coefficient of double reduction in the cultivated tetraploid potato. Theor Appl Genet 85:857–862. https://doi.org/10.1007/BF00225029
Holm S (1979) A simple sequential rejective multiple test procedure. Scand J Stat 6:65–70
Jackson RC, Jackson JW (1996) Gene segregation in autotetraploids: prediction from meiotic configurations. Am J Bot 83:673–678. https://doi.org/10.1002/j.1537-2197.1996.tb12756.x
Jeridi M, Perrier X, Rodier-Goud M, Ferchichi A, D’Hont A, Bakry F (2012) Cytogenetic evidence of mixed disomic and polysomic inheritance in an allotetraploid (AABB) Musa genotype. Ann Bot 110:1593. https://doi.org/10.1093/AOB/MCS220
Kamiri M, Stift M, Srairi I, Costantino G, Moussadik A El, Hmyene A et al (2011) Evidence for non-disomic inheritance in a Citrus interspecific tetraploid somatic hybrid between C. reticulata and C. limon using SSR markers and cytogenetic analysis. Plant Cell Rep 30:1415–1425. https://doi.org/10.1007/s00299-011-1050-x
Kamiri M, Stift M, Costantino G, Dambier D, Kabbage T, Ollitrault P et al (2018) Preferential homologous chromosome pairing in a tetraploid intergeneric somatic hybrid (Citrus reticulata + Poncirus trifoliata) revealed by molecular marker inheritance. Front Plant Sci 9:1–12. https://doi.org/10.3389/fpls.2018.01557
Kijas JMH, Thomas MR, Fowler JCS, Roose ML (1997) Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor Appl Genet 94:701–706. https://doi.org/10.1007/s001220050468
Krug C (1943) Chromosome number in the subfamily Aurantioideae with special reference to the genus Citrus. Bot Gaz 104:602–611
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lee L (1988) Citrus polyploidy—origins and potential for cultivar improvement. Aust J Agric Res 39:735. https://doi.org/10.1071/AR9880735
Mather K (1935) Reductional and equational separation of the chromosomes in bivalents and multivalents. J Genet 30:53–78. https://doi.org/10.1007/BF02982205
Mather K (1936) Segregation and linkage in autotetraploids. J Genet 32:287–314. https://doi.org/10.1007/BF02982683
Muller HJ (1914) A new mode of segregation in Gregory’s tetraploid primulas. Am Nat 48:508–512
Nicolosi E, Deng ZN, Gentile A, La Malfa S, Continella G, Tribulato E (2000) Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet 100:1155–1166. https://doi.org/10.1007/s001220051419
Ollitrault P, Dambier D, Luro F, Froelicher Y (2008) Ploidy manipulation for breeding seedless triploid citrus. Plant breeding reviews. Wiley, Hoboken, pp 323–352
Ollitrault F, Terol J, Pina JA, Navarro L, Talon M, Ollitrault P (2010) Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. Am J Bot 97:e124–e129. https://doi.org/10.3732/ajb.1000280
Ollitrault P, Terol J, Chen C, Federici C, Lotfy S, Hippolyte I et al (2012a) A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics 13:593. https://doi.org/10.1186/1471-2164-13-593
Ollitrault P, Terol J, Garcia-Lor A, Bérard A, Chauveau A, Froelicher Y et al (2012b) SNP mining in C clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics 13:13. https://doi.org/10.1186/1471-2164-13-13
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34:401–437. https://doi.org/10.1146/annurev.genet.34.1.401
Oueslati A, Salhi-Hannachi A, Luro F, Vignes H, Mournet P, Ollitrault P (2017) Genotyping by sequencing reveals the interspecific C. maxima/C. reticulata admixture along the genomes of modern citrus varieties of mandarins, tangors, tangelos, orangelos and grapefruits. PLoS One 12:e0185618. https://doi.org/10.1371/journal.pone.0185618
Perrier X, Jacquemound-Collet JP (2018) DARwin 6.0.18. http://darwin.cirad.fr
Ramsey J (2007) Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae). Heredity 98:143–150. https://doi.org/10.1038/sj.hdy.6800912
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29:467–501. https://doi.org/10.1146/annurev.ecolsys.29.1.467
Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annu Rev Ecol Syst 33:589–639. https://doi.org/10.1146/annurev.ecolsys.33.010802.150437
Ronfort J, Jenczewski E, Bataillon T, Rousset F (1998) Analysis of population structure in autotetraploid species. Genetics 150:921–930
Rouiss H, Bakry F, Froelicher Y, Navarro L, Aleza P, Ollitrault P (2018) Origin of C. latifolia and C. aurantiifolia triploid limes: the preferential disomic inheritance of doubled-diploid “Mexican” lime is consistent with an interploid hybridization hypothesis. Ann Bot 121:571–585. https://doi.org/10.1093/aob/mcx179
Ruiz M, Quiñones A, Martínez-Alcántara B, Aleza P, Morillon R, Navarro L et al (2016) Tetraploidy enhances boron-excess tolerance in carrizo citrange (Citrus sinensis L. Osb. × Poncirus trifoliata L. Raf.). Front Plant Sci 7:1–16. https://doi.org/10.3389/fpls.2016.00701
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 44:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sanford J (1983) Ploidy manipulations. In: Methods in fruit breeding. In: Moore J, Janick J (eds) Purdue University Press, West Lafayettena, pp 100–123
Soltis DE, Segovia-Salcedo MC, Jordon-Thaden I, Majure L, Miles NM, Mavrodiev EV et al (2014) Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytol 202:1105–1117. https://doi.org/10.1111/nph.12756
Starrantino A, Recupero RG (1982) Citrus hybrids obtained in vitro from 2× females × 4× males. In: Proceedings of the International Society of Citriculture, ed. K. Matsumoto (Tokyo, Japan: International Society of Citriculture)
Stebbins GL (1947) Types of polyploids: their classification and significance. Adv Genet 1:403–429. https://doi.org/10.1016/S0065-2660(08)60490-3
Stift M, Berenos C, Kuperus P, van Tienderen PH (2008) Segregation models for disomic, tetrasomic and intermediate inheritance in tetraploids: a general procedure applied to Rorippa (yellow cress) microsatellite data. Genetics 179:2113–2123. https://doi.org/10.1534/genetics.107.085027
Sybenga J (1996) Chromosome pairing affinity and quadrivalent formation in polyploids: do segmental allopolyploids exist? Genome 39:1176–1184. https://doi.org/10.1139/g96-148
Sybenga J (2012) Cytogenetics in genetics and plant breeding. Springer, Berlin, pp 1–5. https://doi.org/10.1007/978-3-642-84083-8_1
Tai GCC (1982a) Estimation of double reduction and genetic parameters in autotetraploids based on 4×–2× and 4×–4× matings. Heredity 49:331–335. https://doi.org/10.1038/hdy.1982.106
Tai GCC (1982b) Estimation of double reduction and genetic parameters of autotetraploids. Heredity 49:63–70. https://doi.org/10.1038/hdy.1982.65
Van de Peer Y, Mizrachi E, Marchal K (2017) The evolutionary significance of polyploidy. Nat Rev Genet 18:411–424. https://doi.org/10.1038/nrg.2017.26
Welch JE (1960) Linkage in Autotetraploid Maize. Genetics 47:367–396
Wu R, Gallo-Meagher M, Littell RC, Zeng ZBB (2001) A general polyploid model for analyzing gene segregation in outcrossing tetraploid species. Genetics 159:869–882
Wu GA, Prochnik S, Jenkins J, Salse J, Hellsten U, Murat F et al (2014) Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol 32:656–662. https://doi.org/10.1038/nbt.2906
Wu GA, Terol J, Ibanez V, López-García A, Pérez-Román E, Borredá C et al (2018) Genomics of the origin and evolution of Citrus. Nature 554:311–316. https://doi.org/10.1038/nature25447
Xu Q, Chen L-L, Ruan X, Chen D, Zhu A, Chen C et al (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45:59–66. https://doi.org/10.1038/ng.2472
Funding
This work was supported by the project RTA2015-00069-00-00 from the Ministry of “Economía y Competividad” and “Instituto Nacional de Investigación y Tecnología Agraria y Agroalimentaria” and the grant ‘Programa de Perfeccionamiento’ resolution 1177/14 from Consejo Directivo of Instituto Nacional de Tecnología Agropecuaria (Argentina).
Author information
Authors and Affiliations
Contributions
PO and PA conceived and designed the experiments. MG performed the experiments. MG, AGL, NO, JC, and PA analyzed the data. PO provided a statistical method for the estimation of PP and maximum DR and new SNP markers from GBS data analysis. MG, JC, and PA wrote the manuscript with input and review of LN and PO. All authors read and approved the final version of this manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by Carlos F. Quiros.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Garavello, M., Cuenca, J., Garcia-Lor, A. et al. Male and female inheritance patterns in tetraploid ‘Moncada’ mandarin. Plant Cell Rep 39, 335–349 (2020). https://doi.org/10.1007/s00299-019-02494-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02494-y