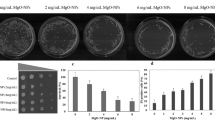
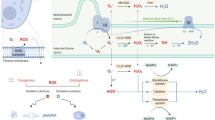
Abstract
The yeast Saccharomyces cerevisiae is capable of responding to various environmental stresses, such as salt stress. Such responses require a complex network and adjustment of the gene expression network. The goal of this study is to further understand the molecular mechanism of salt stress response in yeast, especially the molecular mechanism related to genes BDF1 and HAL2. The Bromodomain Factor 1 (Bdf1p) is a transcriptional regulator, which is part of the basal transcription factor TFIID. Cells lacking Bdf1p are salt sensitive with an abnormal mitochondrial function. We previously reported that the overexpression of HAL2 or deletion of HDA1 lowers the salt sensitivity of bdf1Δ. To better understand the mechanism behind the HAL2-related response to salt stress, we compared three global transcriptional profiles (bdf1Δ vs WT, bdf1Δ + HAL2 vs bdf1Δ, and bdf1Δhda1Δ vs bdf1Δ) in response to salt stress using DNA microarrays. Our results reveal that genes for iron acquisition and cellular and mitochondrial remodeling are induced by HAL2. Overexpression of HAL2 decreases the concentration of nitric oxide. Mitochondrial iron–sulfur cluster (ISC) assembly also decreases in bdf1Δ + HAL2. These changes are similar to the changes of transcriptional profiles induced by iron starvation. Taken together, our data suggest that mitochondrial functions and iron homeostasis play an important role in bdf1Δ-induced salt sensitivity and salt stress response in yeast.




Similar content being viewed by others
References
Almeida B, Buttner S, Ohlmeier S, Silva A, Mesquita A, Sampaio-Marques B, Osorio NS, Kollau A, Mayer B, Leao C, Laranjinha J, Rodrigues F, Madeo F, Ludovico P (2007) NO-mediated apoptosis in yeast. J Cell Sci 120:3279–3288
Bindoli A, Fukuto JM, Forman HJ (2008) Thiol chemistry in peroxidase catalysis and redox signaling. Antioxid Redox Signal 10:1549–1564
Brown GC (2001) Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim Biophys Acta 1504:46–57
Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule J-J, Manon S (2003) The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol Microbiol 47:495–506
Chen L, Liu L, Wang M, Fu J, Zhang Z, Hou J, Bao X (2013) Hal2p functions in Bdf1p-involved salt stress response in Saccharomyces cerevisiae. PLoS One 8:e62110
Chen L, Wang M, Hou J, Liu L, Fu J, Shen Y, Zhang Z, Bao X (2014) Regulation of Saccharomyces cerevisiae MEF1 by Hda1p affects salt resistance of bdf1Δ mutant. FEMS Yeast Res 14:575–585
Chung J, Bachelder RE, Lipscomb EA, Shaw LM, Mercurio AM (2002) Integrin (alpha 6 beta 4) regulation of eIF-4E activity and VEGF translation: a survival mechanism for carcinoma cells. J Cell Biol 158:165–174
Dancis A, Klausner RD, Hinnebusch AG, Barriocanal JG (1990) Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol 10:2294–2301
Deng Y, Guo Y, Watson H, Au WC, Shakoury-Elizeh M, Basrai MA, Bonifacino JS, Philpott CC (2009) Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-Golgi network to the vacuole. J Biol Chem 284:23830–23841
Dumont ME, Ernst JF, Sherman F (1988) Coupling of heme attachment to import of cytochrome-C into yeast mitochondria—studies with heme lyase-deficient mitochondria and altered apocytochromes-C. J Biol Chem 263:15928–15937
Ferreira MCD, Bao XM, Laize V, Hohmann S (2001) Transposon mutagenesis reveals novel loci affecting tolerance to salt stress and growth at low temperature. Curr Genet 40:27–39
Flury F, von Borstel RC, Williamson DH (1976) Mutator activity of petite strains of Saccharomyces cerevisiae. Genetics 83:645–653
Froissard M, Belgareh-Touze N, Dias M, Buisson N, Camadro JM, Haguenauer-Tsapis R, Lesuisse E (2007) Trafficking of siderophore transporters in Saccharomyces cerevisiae and intracellular fate of ferrioxamine B conjugates. Traffic 8:1601–1616
Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360
Grant CM, MacIver FH, Dawes IW (1996) Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet 29:511–515
Hausmann A, Samans B, Lill R, Muhlenhoff U (2008) Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem 283:8318–8330
Ho YH, Gasch AP (2015) Exploiting the yeast stress activated signaling network to inform on stress biology and disease signaling. Curr Genet 61:503–511
Ihrig J, Hausmann A, Hain A, Richter N, Hamza I, Lill R, Muhlenhoff U (2010) Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell 9:460–471
Jones-Carson J, Laughlin J, Hamad MA, Stewart AL, Voskuil MI, Vazquez-Torres A (2008) Inactivation of [Fe–S] metalloproteins mediates nitric oxide-dependent killing of Burkholderia mallei. PLoS One 3:e1976
Kaplan CD, Kaplan J (2009) Iron acquisition and transcriptional regulation. Chem Rev 109:4536–4552
Kaplan J, McVey Ward D, Crisp RJ, Philpott CC (2006) Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta 1763:646–651
Kim D, Yukl ET, Moenne-Loccoz P, Montellano PROd (2006) Fungal heme oxygenases: functional expression and characterization of Hmx1 from Saccharomyces cerevisiae and CaHmx1 from Candida albicans. Biochemistry 45:14772–14780
Lee J-H, Maskos K, Huber R (2009) Structural and functional studies of the yeast class II Hda1 histone deacetylase complex. J Mol Biol 391:744–757
Li L, Miao R, Jia X, Ward DM, Kaplan J (2014) Expression of the yeast cation diffusion facilitators Mmt1 and Mmt2 affects mitochondrial and cellular iron homeostasis: evidence for mitochondrial iron export. J Biol Chem 289:17132–17141
Liu X, Zhang X, Wang C, Liu L, Lei M, Bao X (2007) Genetic and comparative transcriptome analysis of bromodomain factor 1 in the salt stress response of Saccharomyces cerevisiae. Curr Microbiol 54:325–330
Liu X, Yang H, Zhang X, Liu L, Lei M, Zhang Z, Bao X (2009) Bdf1p deletion affects mitochondrial function and causes apoptotic cell death under salt stress. FEMS Yeast Res 9:240–246
Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, Sentenac A, Seraphin B (1994) The yeast Bdf1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res 22:5332–5340
Matangkasombut O, Buratowski S (2003) Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell 11:353–363
Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S (2000) Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Gene Dev 14:951–962
Murguia JR, Belles JM, Serrano R (1995) A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267:232–234
Murguia JR, Belles JM, Serrano R (1996) The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J Biol Chem 271:29029–29033
Pastor MM, Proft M, Pascual-Ahuir A (2009) Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J Biol Chem 284:30307–30317
Philpott CC, Protchenko O (2008) Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell 7:20–27
Protchenko O, Philpott CC (2003) Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J Biol Chem 278:36582–36587
Protchenko O, Ferea T, Rashford J, Tiedeman J, Brown PO, Botstein D, Philpott CC (2001) Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J Biol Chem 276:49244–49250
Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK (2003) Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res 31:3909–3917
Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437–446
Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, Garcia-Arencibia M, Rose C, Luo S, Underwood BR, Kroemer G, O’Kane CJ, Rubinsztein DC (2011) Complex inhibitory effects of nitric oxide on autophagy. Mol Cell 43:19–32
Sawa C, Nedea E, Krogan N, Wada T, Handa H, Greenblatt J, Buratowski S (2004) Bromodomain factor 1 (Bdf1) is phosphorylated by protein kinase CK2. Mol Cell Biol 24:4734–4742
Schneiter R (2007) Intracellular sterol transport in eukaryotes, a connection to mitochondrial function? Biochimie 89:255–259
Singh A, Kaur N, Kosman DJ (2007) The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J Biol Chem 282:28619–28626
Todeschini AL, Condon C, Benard L (2006) Sodium-induced GCN4 expression controls the accumulation of the 5′ to 3′ RNA degradation inhibitor, 3′-phosphoadenosine 5′-phosphate. J Biol Chem 281:3276–3282
Wu J, Carmen AA, Kobayashi R, Suka N, Grunstein M (2001) HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc Natl Acad Sci USA 98:4391–4396
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15
Zhang L, Zhang Y, Zhou Y, An S, Zhou Y, Cheng J (2002) Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J Antimicrob Chemother 49:905–915
Zorov DB, Isaev NK, Plotnikov EY, Zorova LD, Stelmashook EV, Vasileva AK, Arkhangelskaya AA, Khrjapenkova TG (2007) The mitochondrion as janus bifrons. Biochemistry (Mosc) 72:1115–1126
Acknowledgments
This research was supported by the Young Scientists Fund (No. 41401285) of the National Natural Science Foundation of China and the Open Foundation of State Key Laboratory of Estuarine and Coastal Research (SKLEC-KF201412). This work was also supported by the National Natural Science Foundation of China (Nos. 30170021, 30671143, 30570031, 41371257 and 41573071).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
L. Chen and M. Wang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2016_628_MOESM2_ESM.xlsx
Supplementary Table 2. Strains bdf1Δ, bdf1Δ + HAL2, bdf1Δhda1Δ, and WT treated with 0.5 M NaCl for 45 min. The transcription of 1444 genes was significantly changed (ratio >2.0) in bdf1Δ compared to WT. 116 genes were significantly changed (ratio >2.0) in bdf1Δ + HAL2 compared to bdf1Δ. 88 genes were significantly changed (ratio >2.0) compared to bdf1Δ (XLSX 133 kb)
294_2016_628_MOESM3_ESM.docx
Supplementary Table 3. Functional group analysis of the MIPS categories of the transcriptomic data. Comparison of significantly changed genes (≥2 fold) in response to salt treatment (0.5 M NaCl, 45 min) in microarrays for bdf1Δ vs WT (DOCX 15 kb)
294_2016_628_MOESM4_ESM.docx
Supplementary Table 4. Functional group analysis of the MIPS categories of the transcriptomic data. Comparison of significantly changed genes (≥2 fold) in response to salt treatment (0.5 M NaCl, 45 min) in microarrays for bdf1Δ + HAL2 vs bdf1Δ (DOCX 16 kb)
294_2016_628_MOESM5_ESM.docx
Supplementary Table 5. Functional group analysis of the MIPS categories of the transcriptomic data. Comparison of significantly changed genes (≥2 fold) in response to salt treatment (0.5 M NaCl, 45 min) in microarrays for bdf1Δhda1Δ vs bdf1Δ (DOCX 17 kb)
294_2016_628_MOESM6_ESM.docx
Supplementary Table 6. Gene function annotation based on the Saccharomyces Genome Database (the http://www.yeastgenome.org/) shown in the heat map (Fig. 2) in response to salt treatment (0.5 M NaCl, 45 min) in microarrays of bdf1Δ vs WT (DOCX 18 kb)
294_2016_628_MOESM7_ESM.docx
Supplementary Table 7. Gene function annotation based on the Saccharomyces Genome Database (http://www.yeastgenome.org/) shown in the heat map (Fig. 2) in response to salt treatment (0.5 M NaCl, 45 min) in microarrays of (1) bdf1Δ + HAL2 vs bdf1Δ and (2) bdf1Δhda1Δ vs bdf1Δ. (DOCX 23 kb)
Rights and permissions
About this article
Cite this article
Chen, L., Wang, M., Hou, J. et al. HAL2 overexpression induces iron acquisition in bdf1Δ cells and enhances their salt resistance. Curr Genet 63, 229–239 (2017). https://doi.org/10.1007/s00294-016-0628-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0628-9