Abstract
We derive the exact one-step transition probabilities of the number of lineages that are ancestral to a random sample from the current generation of a bi-parental population that is evolving under the discrete Wright–Fisher model with \(n\) diploid individuals. Our model allows for a per-generation recombination probability of \(r\). When \(r=1\), our model is equivalent to Chang’s (Adv Appl Probab 31:1002–1038, 1999) model for the karyotic pedigree. When \(r=0\), our model is equivalent to Kingman’s (Stoch Process Appl 13:235–248, 1982) discrete coalescent model for the cytoplasmic tree or sub-karyotic tree containing a DNA locus that is free of intra-locus recombination. When \(0 < r <1\) our model can be thought to track a sub-karyotic ancestral graph containing a DNA sequence from an autosomal chromosome that has an intra-locus recombination probability \(r\). Thus, our family of models indexed by \(r\in [0,1]\) connects Kingman’s discrete coalescent to Chang’s pedigree in a continuous way as \(r\) goes from \(0\) to \(1\). For large populations, we also study three properties of the ancestral process corresponding to a given \(r\in (0,1)\): the time \(\fancyscript{T}_n\) to a most recent common ancestor (MRCA) of the population, the time \(\fancyscript{U}_n\) at which all individuals are either common ancestors of all present day individuals or ancestral to none of them, and the fraction of individuals that are common ancestors at time \(\fancyscript{U}_n\). These results generalize the three main results of Chang’s (Adv Appl Probab 31:1002–1038, 1999). When we appropriately rescale time and recombination probability by the population size, our model leads to the continuous time Markov chain called the ancestral recombination graph of Hudson (Theor Popul Biol 23:183–201, 1983) and Griffiths (The two-locus ancestral graph, Institute of Mathematical Statistics 100–117, 1991).





Similar content being viewed by others
References
Amato P, Tachibana M, Sparman M, Mitalipov S (2014) Three-parent invitro fertilization: gene replacement for the prevention of inherited mitochondrial diseases. Fertil Steril 101(1):31–35
Athreya K, Ney P (2004) Branching processes. Dover Publications Inc., Mineola, NY, reprint of the 1972 original [Springer, New York)
Barton N, Etheridge A (2011) The relation between reproductive value and genetic contribution. Genetics 188:953–973
Cameron P (1994) Combinatorics: topics, techniques, algorithms. Cambridge University Press, Cambridge
Chang J (1999) Recent common ancestors of all present-day individuals. Adv Appl Probab 31(4):1002–1038 with discussion and reply by the author
Donnelly P, Wiuf C, Hein J, Slatkin M, Ewens W, Kingman J (1999) Discussion: Recent common ancestors of all present-day individuals. Adv Appl Probab 31:1027–1035
Ethier S, Kurtz T (1986) Markov processes: characterization and convergence. Wiley, New York
Fisher R (1930) The Genetical Theory of Natural Selection. Clarenson, Oxford
Gallagher J (2015) UK approves three-person babies. BBC News Retrieved 17 March 2015 from http://www.bbc.com/news/health-31594856a
Gravel S, Steel M (2014) The existence and abundance of ‘ghost’ ancestors in biparental populations. Preprint
Griffiths R (1991) The two-locus ancestral graph. In: Basawa I, Taylor R (eds) Selected Proceedings of the Sheffield Symposium on Applied Probability: Held at the University of Sheffield, Sheffield, August 16–19, 1989, IMS Lecture Notes—onograph Series, vol 18, Institute of Mathematical Statistics, pp 100–117
Griffiths R, Marjoram P (1997) An ancestral recombination graph. In: Donnelly P, Tavaré S (eds) Progress in Population Genetics and Human Evolution, IMA Volumes in Mathematics and its Applications, vol 87. Springer, pp 257–270
Gusfield D (2014) ReCombinatorics: The Algorithmics of Ancestral Recombination Graphs and Explicit Phylogenetic Networks. MIT Press, Cambridge
Hudson R (1983) Properties of a neutral allele model with lntragenic recombination. Theor Popul Biol 23:183–201
Hurst G, Jiggins F (2005) Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc B 272:1525–1534
Kämmerle K (1989) Looking forwards and backwards in a bisexual Moran model. J Appl Probab 26(4):880–885
Kämmerle K (1991) The extinction probability of descendants in bisexual models of fixed population size. J Appl Probab 28(3):489–502
Kemeny J, Snell J (1960) Finite Markov chains. D. van Nostrand Company Inc, Princeton
Kingman J (1982) The Coalescent. Stoch Process Appl 13:235–248
Lachance J (2009) Inbreeding, pedigree size, and the most recent common ancestor of humanity. J Theor Biol 261:238–247
Matsen F, Evans S (2008) To what extant does genealogical ancestry imply genetic ancestry? Theor Pop Biol 74:182–190
Möhle M (1994) Forward and backward processes in bisexual models with fixed population sizes. J Appl Probab 31(2):309–332
Möhle M (1998) A convergence theorem for Markov chains arising in population genetics and the coalescent with selfing. Adv Appl Probab 30:493–512
Ralph P (2009) Most recent common ancestors, genetic inheritance, stochastic gene transcription, and Brownian motion on disconnected sets: a probabilistic analysis of a few models. Ph.D. Thesis
Stein W et al (2009) Sage Mathematics Software (Version 4.2.1). The Sage Development Team, http://www.sagemath.org
Wakeley J, King L, Low B, Ramachandran S (2012) Gene genealogies within a fixed pedigree, and the robustness of Kingman’s coalescent. Genetics 190:1433–1445
White D, Wolff J, Pierson M, Gemmell N (2008) Revealing the hidden complexities of mtDNA inheritance. Mol Ecol 17:4925–4942
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Acknowledgments
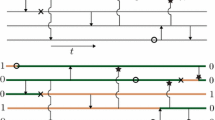
B.T. was partly supported by post-doctoral fellowships at Department of Statistics, University of Oxford, UK (EPSRC grant EP/E05885X/1) and at Instituto de Matemática e Estatística, Universidade de São Paulo, Brasil (CNPq Processo 151782/2010-5 & MaCLinC). B.T. would like to thank Professor Yoshiharu Kohayakawa for many useful discussions related to Theorems 2, 3 and Corollary 1. R.S. was partly supported by a visiting scholarship at Department of Mathematics, Cornell University, Ithaca, NY, USA, a sabbatical grant from College of Engineering, University of Canterbury, and consulting revenues from Wynyard Group, Christchurch, NZ. R.S. thanks Robert C. Griffiths for an introduction to his ARGs, Alison Etheridge for discussions on stationary behaviour of the ancestral size chain, Jae Young Choi and Neil Gemmell for discussions on cytoplasmic inheritance, and Krithika Yogeeswaran for discussions on the nested embeddings in Fig. 3. A.V. was supported by the ANR project MANEGE (ANR-09-BLAN-0215) and R.S. and A.V. were supported in part by the chaire Modélisation Mathématique et Biodiversité of Veolia Environnement–École Polytechnique–Museum National d’Histoire Naturelle–Fondation X. The authors would like to thank the Referees and Associate Editor for their helpful suggestions to improve the presentation of the paper.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A: Galton–Watson processes with Poissonian offspring distribution
In this section, we collect a few facts about Galton–Watson processes. These facts are used in the proofs of Theorems 2 and 3. Recall that a Galton–Watson process with offspring distribution \(\mu \) (a probability distribution on \({\mathbb Z}_+\)) counts the number of individuals alive in each generation in a population evolving as follows: each individual in generation \(k\ge 0\) gives birth to a random number of descendants with law \(\mu \), independently of each other; generation \(k+1\) is then made of all these offspring. If the population becomes extinct at some time, its remains extinct for all later generations (and by extension we say that the Galton–Watson process becomes extinct—stuck at \(0\) —at that time).
The following lemmas summarize Lemmas 4 and 16 of Chang (1999) (with minor modifications, since Chang’s results are for a Galton–Watson process with offspring distribution \(\mathtt {Poisson}(2)\)) and some well-known general results from Chapter 1 of the book by Athreya and Ney (2004). Therefore, we do not give all of their proofs here. We write \(m\) for the expectation of \(\mu \), \(\sigma ^2\) for its variance, and \(\mathbb {P}_i\) (and \(\mathbb {E}_i\)) for the law under which the population starts with \(i\) individuals.
Lemma 11
Let \((Y_t)_{t\in {\mathbb Z}_+}\) be a Galton–Watson process with \(m < 1\). Let \(\tau _0 := \inf \{t : Y_t = 0 \}\) be the extinction time of \((Y_t)_{t\in {\mathbb Z}_+}\). Then for any \(k\in {\mathbb Z}_+\),
Furthermore, if \(\sigma ^2<\infty \) we have for large \(k\)
When \(\mu \) is the law \(\mathtt {Poisson}(\lambda )\) for some \(\lambda >0\), we have \(m=\lambda =\sigma ^2<\infty \).
Lemma 12
Let \((Y_t)_{t\in {\mathbb Z}_+}\) be a Galton–Watson process with offspring distribution \(\mathtt {Poisson}(\lambda )\) for a given \(\lambda >0\). Let \(\psi \) be the probability generating function of \(X\), where \(X\) is a random variable with \(\mathtt {Poisson}(\lambda )\) law. That is, for any \(z\in [0,1]\), \(\psi (z)=\mathbb {E}[z^X]= e^{-\lambda (1-z)}\). Let also \(\varrho \) be the smallest solution in \([0,1]\) to \(\psi (x)=x\). Then
-
(i)
The probability \(\mathbb {P}_1[\tau _0<\infty ]\) that \((Y_t)_{t\in {\mathbb Z}_+}\) becomes extinct in finite time, starting with 1 individual, is equal to \(\varrho >0\).
-
(ii)
The Markov chain \((Y_t/\lambda ^t)_{t\in {\mathbb Z}_+}\) is a martingale. As \(t\) tends to infinity, it converges a.s. to a random variable \(M\) satisfying \(\{M>0\}=\{Y\ \mathrm {survives\ for\, ever}\}\).
-
(iii)
Let \((b_t)_{t\ge 0}\) be a sequence of positive integers such that \(\ln b_t = o(t)\) as \(t \rightarrow \infty \). Then
$$\begin{aligned} \lim _{t \rightarrow \infty } \frac{\ln \mathbb {P}_1[1 \le Y_t \le b_t]}{t} = \ln (\lambda \varrho ). \end{aligned}$$
The first two points in Lemma 12 hold in fact for any Galton–Watson process (with \(\lambda \) replaced by its mean offspring distribution in \((ii)\)). The last point says in essence that, up to polynomial prefactors, the probability that \(Y_t\) is still positive but grows less than exponentially in \(t\) decays like \(e^{-|\ln (\lambda \varrho )|t}\) as \(t\rightarrow \infty \). Observe that the product \(\lambda \varrho \) is always less than one (except in the critical case \(\lambda =1\) in which we shall not be interested), since when \(\lambda <1\) we have \(\varrho =1\), and when \(\lambda >1\), \(\lambda \varrho = \psi '(\varrho )<1\) (\(\psi \) is strictly convex, \(\psi (0)>0\) and \(\varrho <1\) is the smallest positive value at which \(\psi (x)=x\), the largest being \(x=1\)).
Finally, let us give a comparison result between a single family size \((G_t)_{t\in {\mathbb Z}_+}\) (resp., the size of the family of non-descendants \((B_t)_{t\in {\mathbb Z}_+}\)) and a Galton–Watson process with offspring distribution \(\mathtt {Poisson}(1+r)\) (resp., \(\mathtt {Poisson}(1-r)\)).
Lemma 13
Let \((Y^+_t)_{t\in {\mathbb Z}_+}\) (resp., \((Y^-_t)_{t\in {\mathbb Z}_+}\)) be a Galton–Watson process with offspring distribution given by \(\mathtt {Poisson}(1+r)\) (resp., \(\mathtt {Poisson}(1-r)\)). For any \(b>0\), let \(\tau _b^Y:=\inf \{t:\, Y_t\ge b\}\), \(\tau _{0,b}^Y:=\inf \{t:\, Y_t\ge b \hbox { or }Y_t=0\}\) and \(\tau _0^Y:=\inf \{t:\, Y_t=0\}\) (where \(Y=Y^+\) or \(Y^-\)). Define the same quantities for the processes \(G\) and \(B\). Here again, \(\mathbb {P}_i\) denotes the probability measure under which the process under consideration starts at \(i\).
-
(i)
If \(k\) and \(b\) grow with \(n\) in such a way that \(kb^2=o(n)\), then as \(n\rightarrow \infty \)
$$\begin{aligned} \mathbb {P}_1\big [\tau ^G_b>k\big ]= & {} \mathbb {P}_1\big [\tau ^{Y^+}_b>k\big ](1+o(1)) \quad \hbox {and}\quad \mathbb {P}_1\big [\tau ^G_{0,b}>k\big ]\\= & {} \mathbb {P}_1\big [\tau ^{Y^+}_{0,b}>k\big ](1+o(1)). \end{aligned}$$ -
(ii)
If for some \(\alpha \in (0,1/4)\) and \(\gamma \in (2\alpha ,1/2)\) we have \(i=\fancyscript{O}(n^\alpha )\) and \(k=o(n^{1-2\gamma })\), then as \(n\rightarrow \infty \)
$$\begin{aligned} \mathbb {P}_i\big [\tau ^B_0>k\big ]= \mathbb {P}_i\big [\tau ^{Y^-}_0>k\big ](1+o(1)). \end{aligned}$$
Remark 5
Note that \(i=\fancyscript{O}(n^\alpha )\) means that \(i\) is bounded by a constant times \(n^\alpha \), which allows to take \(i\) constant in \(n\) or growing more slowly than \(n^\alpha \).
In words, despite the dependency between the different family sizes in our original model, the early development of a single family \((G_t)_{t\in {\mathbb Z}_+}\) is very close to that of a \(\mathtt {Poisson}(1+r)\) Galton–Watson process. Likewise, as soon as there remains much less than \(n\) individuals in \(B\), the extinction of this subpopulation occurs in the same way as in a \(\mathtt {Poisson}(1-r)\) Galton–Watson process.
Proof
The proof of \((i)\) is similar to that of Lemma 3 of Chang (1999), and so we omit it here. The proof of \((ii)\) follows the same lines but is a bit more complex. Our main aim is to show that as long as the processes do not grow too much and we do not look at too many generations, their transition probabilities are equivalent. Then, since \(Y^-\) starting below \(n^\alpha \) will not grow beyond \(n^\gamma \) before going extinct with very high probability, neither will \(B\) and extinction will occur roughly at the same time (in distribution) for both.
Let us thus consider \(x,y\le n^\gamma \). Recall that conditionally on \(B_t=x\), \(B_{t+1}\sim \mathtt {Bin}(n,(1-r)\frac{x}{n} + r\frac{x^2}{n^2})\). Since the sum of \(x\) independent \(\mathtt {Poisson}(1-r)\) random variables has the law \(\mathtt {Poisson}((1-r)x)\), we have for any \(t\in {\mathbb Z}_+\)
But \(x,y\le n^\gamma \ll n\), and so a first order Taylor expansion gives us that
and
Together with the fact that \(n!/((n-y)!\,n^y)\le 1\), we obtain that
for a constant \(C_r>0\) independent of \(x\) and \(y\) (recall that \(\gamma <1/2\)).
The same analysis, separating the cases \(y=0,1\) and \(y\ge 2\) and using the fact that for \(1< y\le n^\gamma \) we have
shows that
for a constant \(C'_r\) independent of \(x\) and \(y\). Putting together Eqs. (8.5) and (8.6), we obtain that
where the remainder is uniform in \(x\in \{1,\ldots ,n^\gamma \}\) and \(y\in \{0,\ldots ,n^\gamma \}\). As a consequence, for any \(x_0,\ldots ,x_{k-1}\in \{1,\ldots ,n^\gamma \}\) and \(x_k\in \{0,\ldots ,n^\gamma \}\), we have
Summing over all paths corresponding to the event considered and using the fact that \(kn^{2\gamma -1}=o(1)\) as \(n\rightarrow \infty \), we can write that
and
the latter being the probabilities that the process leaves \(\{1,\ldots ,n^\gamma -1\}\) before time \(k\) and by going over \(n^\gamma \). Now,
From the above, the first term in the r.h.s. is equal to the corresponding term for \(Y^-\) up to a vanishing error term. As concerns the second quantity in the r.h.s., it is bounded by
To finish the proof, let us show that the probability that \(Y^-\), starting below \(n^\alpha \), reaches \(n^\gamma \) before becoming extinct tends to \(0\) as \(n\rightarrow \infty \). Together with Eq. (8.7), this will give us the desired result since Eq. (8.7) holds also with \(B\) replaced by \(Y^-\).
Since \(Y^-\) cannot grow beyond \(n^\gamma \) unless one of the \(i\le C n^\alpha \) families emanating from an initial individual reaches \(n^{\gamma -\alpha }/C\), we have
But \(\mathbb {E}_1[Y^-_j]=(1-r)^j\), and so the Markov inequality applied to each term in the sum gives us
As \(\gamma >2\alpha \), this quantity goes to zero as \(n\) tends to infinity.
Appenndix B: Proof of Eq. (8.1)
Here we derive the approximation of Eq. (8.1) in detail.
Note first that we have the following approximation:
We will first consider some special cases. Fix \(I,J,K\) as before such that \(|I| = i, |K| = k, |J| = j\). For \(M\subseteq I\), let \(A(M)\) be the set of parents of vertices in \(M\).
Lemma 14
Proof
We have \(|I| = i, |K| = s, |J| = i+s\). In this case, no two vertices in \(I\) have a common parent, hence
The first factor is the number of ways to select \(A(K)\). The second factor is the number of ways to assign parents to vertices in \(K\), each vertex being assigned 2 distinct parents. The last factor is the number of ways to assign parents to vertices in \(I{\backslash }K\), each vertex being assigned a distinct parent.
Lemma 15
Proof
We have 3 cases.
Case 1 \(|A(K)| = 2s+2\), \(|A(I \backslash K)| = i - s - 2\), and \(A(I \backslash K) = J \backslash A(K)\). The contribution to \(|B(i+s|i,s+1)|\) in this case is
The first factor is the number of ways to select \(A(K)\). The second factor is the number of ways to assign parents to vertices in \(K\), each vertex being assigned 2 distinct parents. The last two factors together give the number of ways to assign parents to vertices in \(I\backslash K\); which is the number of onto maps from \(A(I\backslash K)\) to \(J \backslash A(K)\).
Case 2 \(|A(K)| = 2s+2\), \(|A(I \backslash K)| = i - s - 1\), and \(|A(K) \cap A(I \backslash K)| = 1\). The contribution to \(|B(i+s|i,s+1)|\) in this case is
The first two factors are as in Case 1. The third factor is the number of ways to select the (only) vertex in \(A(K) \cap A(I \backslash K)\). The last factor is the number of ways to assign distinct parents to vertices in \(I\backslash K\).
Case 3 \(|A(K)| = 2s+1\) and \(A(I \backslash K) = J \backslash A(K)\). We have \(|A(I \backslash K)| = i - s - 1\). Also, there are \(s-1\) vertices in \(K\) with two distinct parents each, 2 vertices in \(K\) with one common parent, and \(i-s-1\) vertices in \(I\backslash K\) with distinct parents. The contribution to \(|B(i+s|i,s+1)|\) in this case is
The first factor gives the number of partitions of \(J\) in 3 parts as described above. The second factor is the number of ways to select the two vertices in \(K\) that have a common parent; and the third factor is the number of ways to assign 2 parents to each of them, with one parent in common. The forth factor is the number of ways to assign parents to the remaining \(s-1\) vertices in \(K\), each vertex being assigned 2 distinct parents. The last factor is the number of ways to assign distinct parents to vertices in \(I\backslash K\).
Now \(|B(i+s,i,s+1)|\) is obtained by adding the contributions in Eqs. (8.10), (8.11) and (8.12).
Let us now return to the Proof of Eq. (8.1) and consider the case \(j\ge i\). We must have \(k \ge j-i\), i.e., more recombinants than additional lineages. To find an approximation of \(^{n,r}P_{i,j}\), we use the expression obtained in Theorem 1 [more precisely, we use Eq. (4.4)]. We first evaluate the order of magnitude (in \(n\)) of each term appearing in the sum over \(k\in \{j-i,\ldots ,i\}\). We have
Hence, the term corresponding to \(k\) will be of the order of \(\fancyscript{O}(n^{-2})\) whenever \(k\ge j-i+2\). From now on, we thus consider the terms \(k=j-i\) and \(k=j-i+1\) only. Let us write \(j=i+s\), with \(0\le s\le i\). Suppose first that \(k=s\). Using again the notation \(|B(j|i,k)|\) for the number of bipartite graphs defined in Sect. 4 (where we replaced the sets \(I\), \(J\), \(K\) by their cardinalities since only these quantities matter), we can write
where the first equality uses Eqs. (8.13) and (8.8).
Let us now suppose that \(k=s+1\). This time, we have
where we have used Eq. (8.9). Combining the above, we obtain the desired approximation when \(j\ge i\).
Next, we consider the case where \(j = i-s\), with \(s > 0\). Using again Eq. (8.13), we see that the terms appearing in the sum over \(k\) in the expression of \(^{n,r}P_{i,j}\) will be \(\fancyscript{O}(1/n^2)\) whenever \(s+k\ge 2\), which will be the case whenever \(k\ge 1\) or \(k=0\) and \(s\ge 2\). We thus concentrate on the case \(k=0\) and \(s=1\) only, corresponding to the scenario where a single pair of lineages coalesces and no recombinations occur. Since there are \(\left( {\begin{array}{c}i\\ 2\end{array}}\right) \) possible choices for the pair of lineages that coalesces and then \((i-1)!\) possible allocations of parents, we have \(|B(i-1|i,0)| = (i-1)!\left( {\begin{array}{c}i\\ 2\end{array}}\right) \) and thus
This entails the approximation given on the first line of Eq. (8.1).
Rights and permissions
About this article
Cite this article
Sainudiin, R., Thatte, B. & Véber, A. Ancestries of a recombining diploid population. J. Math. Biol. 72, 363–408 (2016). https://doi.org/10.1007/s00285-015-0886-z
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-015-0886-z
Keywords
- Kingman’s coalescent
- Chang’s pedigree
- Griffiths’ and Hudson’s ancestral recombination graphs
- Zygotic/karyotic/cytoplasmic/sub-karyotic ancestral graphs