Abstract.
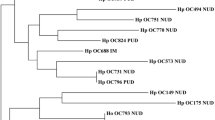
Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori vary, particularly in their mid region (which may be type m1 or m2) and their signal peptide coding region (type s1 or s2). We investigated nucleotide diversity among vacA alleles in strains from several locales in Asia, South America, and the USA. Phylogenetic analysis of vacA mid region sequences from 18 strains validated the division into two main groups (m1 and m2) and showed further significant divisions within these groups. Informative site analysis demonstrated one example of recombination between m1 and m2 alleles, and several examples of recombination among alleles within these groups. Recombination was not sufficiently extensive to destroy phylogenetic structure entirely. Synonymous nucleotide substitution rates were markedly different between regions of vacA, suggesting different evolutionary divergence times and implying horizontal transfer of genetic elements within vacA. Non-synonymous/synonymous rate ratios were greater between m1 and m2 sequences than among m1 sequences, consistent with m1 and m2 alleles encoding functions fitting strains for slightly different ecological niches.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 9 March 1999 / Accepted: 18 June 1999
Rights and permissions
About this article
Cite this article
Atherton, J., Sharp, P., Cover, T. et al. Vacuolating Cytotoxin (vacA) Alleles of Helicobacter pylori Comprise Two Geographically Widespread Types, m1 and m2, and Have Evolved Through Limited Recombination. Curr Microbiol 39, 211–218 (1999). https://doi.org/10.1007/s002849900447
Issue Date:
DOI: https://doi.org/10.1007/s002849900447