Abstract
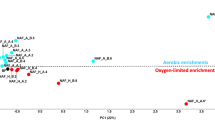
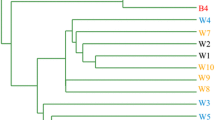
The stimulation of groundwater bacteria to form biofilms, for the remediation of polluted aquifers, is subjected to environmental regulations that included measurement of effects on microbial biodiversity. Groundwater microorganisms contain a proportion of unidentified and uncharacterized ultramicrobacteria (UMB) that might play a major role in the bioclogging of geological materials. This study aimed to assess the changes in genetic and metabolic biodiversity when a community of UMB, isolated from groundwater, is stimulated to form biofilms on a ceramic surface. UMB were stimulated with aerobic conditions and injection of molasses, in reactors reproducing groundwater composition and temperature. Concentration of planktonic viable UMB, secretion of extracellular polymeric substances (EPS), and biofilm thickness were monitored. The assessment of changes in biodiversity was achieved by comparing the initial UMB community to the biofilm community, using the single strand conformational polymorphism (SSCP) method, the cloning and sequencing of 16S rRNA gene (16S rDNA) sequences, and the Biolog microplate system. The hypothesis stating that indigenous UMB would play a significant role of in the biofilm development was corroborated. Within 13 days of stimulation, the UMB produced 700 mg L−1 of planktonic EPS and formed a biofilm up to a thickness of 1100 μm. This stimulation led to a decrease in genetic diversity and an increase in metabolic diversity. The decrease in genetic diversity was shown by a reduced number of single strand DNA fragments in the SSCP profiles. As such, 16S rDNA sequences from the biofilm revealed the predominance of four bacterial groups: Zoogloea, Bacillus/Paenibacillus, Enterobacteriaceae, and Pseudomonads. A significant increase in metabolic diversity was shown by a highest substrate richness profile and a lower substrate evenness profile of the biofilm bacterial population (p=0.0 and p=0.09, respectively). This higher metabolic diversity might be a consequence of the stimulation that seemed to favor the growth of bacteria having a high nutritional versatility. Stimulation of UMB, isolated from groundwater, was effective to form a biofilm having a high metabolic biodiversity. This combination of molecular-based and metabolic-based methods expanded the insight into monitoring the changes in bacterial biodiversity.
Similar content being viewed by others
References
Gavaskar AR, Gupta N, Sass BM, Janosy RJ, O'Sullivan D (1998) Permeable Barriers for Groundwater Remediation—Design, Construction, and Monitoring. Battelle Press, Columbus, OH, pp 1–176
Bellamy KL, de Lint N, Cullimore DR, Abiola A (1993) In-Situ Intercedent Biological Barriers for the Containment and Remediation of Contaminated Groundwater. Droycon Bioconcepts Inc, Regina, SK, Canada, pp
Cunningham AB, Characklis WG, Abedeen F, Crawford D (1991) Influence of biofilm accumulation on porous media hydrodynamics. Environ Sci Technol 25:1305–1311
Vandevivere P, Baveye P (1992) Relationship between transport of bacteria and their clogging efficiency in sand columns. Appl Environ Microbiol 58:2523–2530
Sharp RR, Gerlach R, Cunningham A (1999) Bacterial transport issues related to subsurface biobarriers. In: In Situ and On-Site Bioremediation, the Fifth International Symposium. Battelle Press, San Diego
Morita RY (1982) Starvation-survival of heterotrophs in the marine environment. In: Marshall KC (ed) Advances in Microbial Ecology, vol 6. Plenum Press, New York, pp 171–193
Iizuka T, Yamanaka S, Nishiyama T, Hiraishi A (1998) Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J Gen Appl Microbiol 44:75–84
Kemp PF, Sherr BF, Sherr EB, Cole JJ (1993) Handbook of Methods in Aquatic Microbial Ecology. Lewis Publishers, Boca Raton, FL, pp 1–777
Janssen PH, Schuhmann A, Morschel E, Rainey FA (1997) Novel aerobic ultramicrobacteria belonging to the verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol 63:1382–1388
Johnston CD, Trefry MG, Rayner JL, Ragusa SR, De Zoysa DS, Davis GB (1999) In situ bioclogging for the confinement and remediation of groundwater hydrocarbon plumes. In: Proceedings of the 1999 Contaminated Site Remediation Conference—Contaminated Site Remediation: Challenges Posed by Urban and Industrial Contaminants, March 21–25, Fremantle, Western Australia
Environment Canada (1997) New Substances Notification Regulations. In: Canadian Environmental Protection Act (CEPA) (available at: http://www.ec.gc.ca/CEPARegistry/regulations/DetailReg.cfm)
United States Environmental Protection Agency (1997) Point to consider in the preparation of TSCA biotechnology submissions for microorganisms. In: Toxic Substances Control Act (TSCA) Biotechnology Program, Office of Pollution Prevention and Toxics, Washington, DC, p 71 (available at: http://www.epa.gov/opptintr/biotech/index.html)
Ketcheson DR, Zwiers WG (1997) Evaluation of in-situ restoration technologies for a DNAPL impacted fractured carbonate rock site at Smithville, Ontario. In: Air and Waste Management Association's 90th Annual Meeting and Exhibition, Toronto, Ontario, Canada
Schut F, Gottschal JC, Prins RA (1997) Isolation and characterization of the marine ultramicrobacterium Sphingomonas sp. Strain RB2256. FEMS Microbiol Rev 20:363–369
Novitsky JA, Morita RY (1976) Morphological characterization of small cells resulting from nutrient starvation of a psychrophilic marine Vibrio. Appl Environ Microbiol 32:617–622
Power M, van der Meer JR, Tchelet R, Egli T, Eggen R (1998) Molecular-based methods can contribute to assessment of toxicological risks and bioremediation strategies. J Microbiol Methods 32:107–119
White DC, Flemming CA, Leung KT, Macnaughton SJ (1998) In situ microbial ecology for quantitative appraisal, monitoring and risk assessment of pollution remediation in soils, the subsurface, the rhizosphere and in biofilms. J Microbiol Methods 32:93–105
Lehman RM, Colwell FS, Ringelberg DB, White DC (1995) Combined microbial community-level analyses for quality assurance of terrestrial subsurface cores. J Microbiol Methods 22:263–281
El Fantroussi S, Verschuere L, Verstraete W, Top EM (1999) Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol 65:982–988
Head IM, Saunders JR, Pickup RW (1998) Microbial evoluation, diversity, and ecology: A decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol 35:1–21
Marilley L, Vogt G, Blanc M, Aragno M (1998) Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis. Plant Soil 198:219–224
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Konopka A, Oliver L, Turco RFJ (1998) The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb Ecol 35:103–115
Ross N, Deschênes L, Bureau J, Clément B, Comeau Y, Samson R (1998) Ecotoxicological assessment and effects of physicochemical factors on biofilm development in groundwater conditions. Environ Sci Technol 32:1105–1111
Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R (1999) Live/dead Baclight: Application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. Microbiol Methods 37:77–86
Hacking AJ, Taylor IWF, Jarman TR, Govan JRW (1983) Alginate biosynthesis by Pseudomonas mendocina. J Gen Microbiol 129:3473–3480
Characklis WG, Marshall KC (1990) Biofilms. John Wiley & Sons, Inc, New York, pp 1–796
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Biology a Laboratory Manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp
Lee DH, Zo YG, Kim SJ (1996) Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation-polymorphism. Appl Environ Microbiol 62:3112–3120
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA 69:1408–1412
Felsenstein J (1989) Phylip: Phylogeny inference package (version 3.2). Cladistics 5:164–166
Kimura M (1980) A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evolution 16:111–120
Mathieu L, Paquin JL, Block JC, Randon G, Maillard J, Reasoner D (1992) Paramètres gouvernant la prolifération bacterienne dans les réseaux de distribution. Rev sci eau 5:91–112
Zak JC, Willig MR, Moorhead DL, Wildman HG (1994) Functional diversity of microbial communities: A quantitative approach. Soil Biol Biochem 26:1101–1108
Janssen PH, Schuhmann A, Bak F, Liesak W (1996) Disproportionation of inorganic sulfur compounds by the sulfate-reducing bacterium Desulfocapsa thiozymogenes ge. Nov. Sp. Nov Arch Microbiol 166:184–192
Rosado AS, Duarte GF, Seldin L, Van Elsas JD (1998) Genetic diversity of nifh gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl Environ Microbiol 64:2770–2779
Brenner DJ, Muller HE, Steigerwalt AG, Whitney AM, O'Hara CM, Kampfer P (1998) Two new Rahnella genemospecies that cannot be phenotypically differentiated from Rahnella aquatilis. Int J Syst Bacteriol 48:141–149
Bos R, van der Mei HC, Busscher HJ (1999) Physicochemistry of initial microbial adhesive interactions—its mechanisms and methods for study. FEMS Microbiol Rev 23:179–230
Mueller RF, Characklis WG, Jones WL, Sears JT (1992) Characterization of initial events in bacterial surface colonization by two Pseudomonas species using image analysis. Biotechnol Bioeng 39:1161–1170
Bergey DH (1984) Bergey's Manual of Systematic Bacteriology, J.G. Holt, pp 1–1599
Haack SK, Garchow H, Klug MJ, Forney LJ (1995) Analysis of factors affecting the accuracy, reproducibility, and interpretation of microbial community carbon source utilization patterns. Appl Environ Microbiol 61:1458–1468
Smalla K, Wachtendorf U, Heuer H, Liu W-T, Forney L (1998) Analysis of Biolog GN substrate utilization patterns by microbial communities. Appl Environ Microbiol 64:1220–1225
Verschuere L, Fievez V, Van Vooren L, Verstraete W (1997) The contribution of individual populations to the Biolog pattern of model microbial communities. FEMS Microbiol Ecol 24:353–362
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem 28:213–221
Howard PJA (1997) Analysis of data from Biolog plates: Comments on the method of Garland and Mills. Soil Biol Biochem 29:1755–1757
Fuller ME, Scow KM, Lau S, Ferris H (1997) Trichloroethylene (TCE) and toluene effects on the structure and function of the soil community. Soil Biol Biochem 29:75–89
Talpsep E, Heinaru E, Truu J, Laht T, Heinaru A, Wand H, Stottmeister U (1997) Functional dynamics of microbial populations in waters contaminated with phenolic leachate. Oil Shale 14:435–453
Bääth E, Diaz-Ravina M, Frostegard A, Campbell CD (1998) Effect of metal-rich sludge amendments on the soil microbial community. Appl Environ Microbiol 64:238–245
Marshall KC (1984) Advances in Microbial Ecology, Vol 7. Plenum Press, New York, pp 1–223
Horowitz A, Krichevsky MI, Atlas RM (1983) Characteristics and diversity of subarctic marine oligotrophic, stenoheterotrophic, and euryheterotrophic bacterial populations. Can J Microbiol 29:527–535
Stocker JG (1968) The ecology of a diatom community in a thermal stream. Br Phycol Bul 3:501–514
Derry AM, Staddon WJ, Trevors JT (1998) Functional diversity and community structure of micro-organisms of uncontaminated and creosote-contaminated soils as determined by sole-carbon-source-utilization. World J Microbiol Biotechnol 14:571–578
Cusack F, Singh S, McCarthy C, Grieco J, De Rocco M, Ngyen D, Lappin-Scott H, Costerton JW (1992) Enhanced oil recovery—three-dimensional sandpack simulation of ultramicrobacteria resuscitation in reservoir formation. J Gen Microbiol 138:647–655
Egli T (1995) The ecological and physiological significance of the growth of heterotrophic micro-organisms with mixtures of substrates. FEMS Microbiol Ecol 20:363–369
VanDemark PJ, Batzing BL (1987) The Microbes—an Introduction to Their Nature and Importance. Benjamin/Cummings Science, Inc, San Francisco, pp 1–991
Harder J, Probian C (1997) Anaerobic mineralisation of cholesterol by a novel type of denitrifying bacterium. Arch Microbiol 167:269–274
Rossello-Mora RA, Wagner M, Amann R, Schleifer KH (1995) The abundance of Zoogloea ramigera in sewage treatment plants. Appl Environ Microbiol 61:702–707
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298
Marshall BJ, Ohye DF (1966) Bacillus macquariensis n.Sp., a psychrotrophic bacterium from sub-antarctic soil. J Gen Microbiol 44:41–46
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ross, N., Villemur, R., Marcandella, É. et al. Assessment of changes in biodiversity when a community of ultramicrobacteria isolated from groundwater is stimulated to form a biofilm. Microb Ecol 42, 56–68 (2001). https://doi.org/10.1007/s002480000085
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s002480000085