Abstract
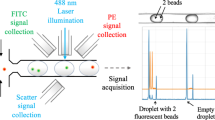
The use of high-throughput multiplexed screening platforms has attracted significant interest in the field of on-site disease detection and diagnostics for their capability to simultaneously interrogate single-cell responses across different populations. However, many of the current approaches are limited by the spectral overlap between tracking materials (e.g., organic dyes) and commonly used fluorophores/biochemical stains, thus restraining their applications in multiplexed studies. This work demonstrates that the downconversion emission spectra offered by rare earth (RE)-doped β-hexagonal NaYF4 nanoparticles (NPs) can be exploited to address this spectral overlap issue. Compared to organic dyes and other tracking materials where the excitation and emission is separated by tens of nanometers, RE elements have a large gap between excitation and emission which results in their spectral independence from the organic dyes. As a proof of concept, two differently doped NaYF4 NPs (europium: Eu3+, and terbium: Tb3+) were employed on a fluorescent microscopy-based droplet microfluidic trapping array to test their feasibility as spectrally independent droplet trackers. The luminescence tracking properties of Eu3+-doped (red emission) and Tb3+-doped (green emission) NPs were successfully characterized by co-encapsulating with genetically modified cancer cell lines expressing green or red fluorescent proteins (GFP and RFP) in addition to a mixed population of live and dead cells stained with ethidium homodimer. Detailed quantification of the luminescent and fluorescent signals was performed to confirm no overlap between each of the NPs and between NPs and cells. Thus, the spectral independence of Eu3+-doped and Tb3+-doped NPs with each other and with common fluorophores highlights the potential application of this novel technique in multiplexed systems, where many such luminescent NPs (other doped and co-doped NPs) can be used to simultaneously track different input conditions on the same platform.

ᅟ








Similar content being viewed by others
References
Saadatpour A, Lai S, Guo G, Yuan G-C. Single-cell analysis in cancer genomics. Trends Genet. 2015;31(10):576–86. https://doi.org/10.1016/j.tig.2015.07.003.
Tellez-Gabriel M, Ory B, Lamoureux F, Heymann M-F, Heymann D. Tumour heterogeneity: the key advantages of single-cell analysis. Int J Mol Sci. 2016;17(12):2142. https://doi.org/10.3390/ijms17122142.
Tsoucas D, Yuan G-C. Recent progress in single-cell cancer genomics. Curr Opin Genet Dev. 2017;42:22–32. https://doi.org/10.1016/j.gde.2017.01.002.
Schneider T, Kreutz J, Chiu DT. The potential impact of droplet microfluidics in biology. Anal Chem. 2013;85(7):3476–82. https://doi.org/10.1021/ac400257c.
Shang L, Cheng Y, Zhao Y. Emerging droplet microfluidics. Chem Rev. 2017;117(12):7964–8040. https://doi.org/10.1021/acs.chemrev.6b00848.
Newell EW, Cheng Y. Mass cytometry: blessed with the curse of dimensionality. Nat Immunol. 2016;17:890. https://doi.org/10.1038/ni.3485.
O'Donnell EA, Ernst DN, Hingorani R. Multiparameter flow cytometry: advances in high resolution analysis. Immune Netw. 2013;13(2):43–54. https://doi.org/10.4110/in.2013.13.2.43.
Wyatt Shields Iv C, Reyes CD, Lopez GP. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip. 2015;15(5):1230–49. https://doi.org/10.1039/C4LC01246A.
Hu P, Zhang W, Xin H, Deng G. Single cell isolation and analysis. Front Cell Dev Biol. 2016;4:116. https://doi.org/10.3389/fcell.2016.00116.
Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165(4):780–91. https://doi.org/10.1016/j.cell.2016.04.019.
Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nat Immunol. 2014;15(2):128–35. https://doi.org/10.1038/ni.2796.
Zhou Y, Basu S, Wohlfahrt KJ, Lee SF, Klenerman D, Laue ED, et al. A microfluidic platform for trapping, releasing and super-resolution imaging of single cells. Sensors Actuator B Chem. 2016;232:680–91. https://doi.org/10.1016/j.snb.2016.03.131.
Holton AB, Sinatra FL, Kreahling J, Conway AJ, Landis DA, Altiok S. Microfluidic biopsy trapping device for the real-time monitoring of tumor microenvironment. PLoS One. 2017;12(1):e0169797. https://doi.org/10.1371/journal.pone.0169797.
Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc. 2017;12(1):44–73. https://doi.org/10.1038/nprot.2016.154.
Ma J, Zhan L, Li RS, Gao PF, Huang CZ. Color-encoded assays for the simultaneous quantification of dual cancer biomarkers. Anal Chem. 2017;89(16):8484–9. https://doi.org/10.1021/acs.analchem.7b02033.
Lan F, Haliburton JR, Yuan A, Abate AR. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat Commun. 2016;7:11784. https://doi.org/10.1038/ncomms11784.https://www.nature.com/articles/ncomms11784#supplementary-information.
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–201. https://doi.org/10.1016/j.cell.2015.04.044.
Chen C-H, Miller MA, Sarkar A, Beste MT, Isaacson KB, Lauffenburger DA, et al. Multiplexed protease activity assay for low volume clinical samples using droplet based microfluidics and its application to endometriosis. J Am Chem Soc. 2013;135(5):1645–8. https://doi.org/10.1021/ja307866z.
Rane TD, Zec HC, Wang TH. A barcode-free combinatorial screening platform for matrix metalloproteinase screening. Anal Chem. 2015;87(3):1950–6. https://doi.org/10.1021/ac504330x.
Fan J, Hu M, Zhan P, Peng X. Energy transfer cassettes based on organic fluorophores: construction and applications in ratiometric sensing. Chem Soc Rev. 2013;42(1):29–43. https://doi.org/10.1039/C2CS35273G.
Abdel-Mottaleb MMA, Beduneau A, Pellequer Y, Lamprecht A. Stability of fluorescent labels in PLGA polymeric nanoparticles: quantum dots versus organic dyes. Int J Pharm. 2015;494(1):471–8. https://doi.org/10.1016/j.ijpharm.2015.08.050.
Montón H, Nogués C, Rossinyol E, Castell O, Roldán M. QDs versus Alexa: reality of promising tools for immunocytochemistry. J Nanobiotechnol. 2009;7(1):4. https://doi.org/10.1186/1477-3155-7-4.
Wen C-Y, Xie H-Y, Zhang Z-L, Wu L-L, Hu J, Tang M, et al. Fluorescent/magnetic micro/nano-spheres based on quantum dots and/or magnetic nanoparticles: preparation, properties, and their applications in cancer studies. Nanoscale. 2016;8(25):12406–29. https://doi.org/10.1039/C5NR08534A.
Leng Y, Wu W, Li L, Lin K, Sun K, Chen X, et al. Magnetic/fluorescent barcodes based on cadmium-free near-infrared-emitting quantum dots for multiplexed detection. Adv Funct Mater. 2016;26(42):7581–9. https://doi.org/10.1002/adfm.201602900.
Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969. https://doi.org/10.1038/nbt994.https://www.nature.com/articles/nbt994#supplementary-information.
Duan N, Wu S, Yu Y, Ma X, Xia Y, Chen X, et al. A dual-color flow cytometry protocol for the simultaneous detection of Vibrio parahaemolyticus and Salmonella typhimurium using aptamer conjugated quantum dots as labels. Anal Chim Acta. 2013;804:151–8. https://doi.org/10.1016/j.aca.2013.09.047.
Buranda T, Wu Y, Sklar LA. Quantum dots for quantitative flow cytometry. Methods Mol Biol. 2011;699:67–84. https://doi.org/10.1007/978-1-61737-950-5_4.
Ghrera AS, Pandey CM, Ali MA, Malhotra BD. Quantum dot-based microfluidic biosensor for cancer detection. Appl Phys Lett. 2015;106(19):193703. https://doi.org/10.1063/1.4921203.
Wegner KD, Hildebrandt N. Quantum dots: bright and versatile in vitro and in vivo fluorescence imaging biosensors. Chem Soc Rev. 2015;44(14):4792–834. https://doi.org/10.1039/C4CS00532E.
Gai SL, Li CX, Yang PP, Lin J. Recent progress in rare earth micro/nanocrystals: soft chemical synthesis, luminescent properties, and biomedical applications. Chem Rev. 2014;114(4):2343–89. https://doi.org/10.1021/cr4001594.
Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5:763. https://doi.org/10.1038/nmeth.1248.https://www.nature.com/articles/nmeth.1248#supplementary-information.
Dong H, Du SR, Zheng XY, Lyu GM, Sun LD, Li LD, et al. Lanthanide nanoparticles: from design toward bioimaging and therapy. Chem Rev. 2015;115(19):10725–815. https://doi.org/10.1021/acs.chemrev.5b00091.
Haase M, Schafer H. Upconverting nanoparticles. Angew Chem Int Ed. 2011;50(26):5808–29. https://doi.org/10.1002/anie.201005159.
Eliseeva SV, Bunzli JCG. Lanthanide luminescence for functional materials and bio-sciences. Chem Soc Rev. 2010;39(1):189–227. https://doi.org/10.1039/b905604c.
Oliveira E, Bértolo E, Núñez C, Pilla V, Santos HM, Fernández-Lodeiro J, et al. Green and red fluorescent dyes for translational applications in imaging and sensing analytes: a dual-color flag. ChemistryOpen. 2018;7(1):9–52. https://doi.org/10.1002/open.201700135.
Lee H, Park HJ, Park C-S, Oh E-T, Choi B-H, Williams B, et al. Response of breast cancer cells and cancer stem cells to metformin and hyperthermia alone or combined. PLoS One. 2014;9(2):e87979. https://doi.org/10.1371/journal.pone.0087979.
Zhou J, Liu Q, Feng W, Sun Y, Li F. Upconversion luminescent materials: advances and applications. Chem Rev. 2015;115(1):395–465. https://doi.org/10.1021/cr400478f.
Chen G, Qiu H, Prasad PN, Chen X. Upconversion nanoparticles: design, nanochemistry, and applications in theranostics. Chem Rev. 2014;114(10):5161–214. https://doi.org/10.1021/cr400425h.
Li CX, Yang J, Quan ZW, Yang PP, Kong DY, Lin J. Different microstructures of ss-NaYF4 fabricated by hydrothermal process: effects of pH values and fluoride sources. Chem Mater. 2007;19(20):4933–42. https://doi.org/10.1021/cm071668g.
Wang F, Liu XG. Recent advances in the chemistry of lanthanide-doped upconversion nanocrystals. Chem Soc Rev. 2009;38(4):976–89. https://doi.org/10.1039/b809132n.
Dorman JA, Choi JH, Kuzmanich G, Bargar JR, Chang JP. Optimizing the crystal environment through extended x-ray absorption fine structure to increase the luminescent lifetimes of Er3+ doped Y2O3 nanoparticles. J Appl Phys. 2012;111(8). https://doi.org/10.1063/1.3702789.
Sabnis A, Rahimi M, Chapman C, Nguyen KT. Cytocompatibility studies of an in situ photopolymerized thermoresponsive hydrogel nanoparticle system using human aortic smooth muscle cells. J Biomed Mater Res Part B Appl Biomater. 2009;91(1):52–9. https://doi.org/10.1002/jbm.a.32194.
Wong DY, Ranganath T, Kasko AM. Low-dose, long-wave UV light does not affect gene expression of human mesenchymal stem cells. PLoS One. 2015;10(9):e0139307. https://doi.org/10.1371/journal.pone.0139307.
Naczynski DJ, Tan MC, Zevon M, Wall B, Kohl J, Kulesa A, et al. Rare-earth-doped biological composites as in vivo shortwave infrared reporters. Nat Commun. 2013;4:2199. https://doi.org/10.1038/ncomms3199.
Yang NJ, Hinner MJ. Getting across the cell membrane: an overview for small molecules, peptides, and proteins. Methods Mol Bio. 2015;1266:29–53. https://doi.org/10.1007/978-1-4939-2272-7_3.
Wu P, Yan X-P. Doped quantum dots for chemo/biosensing and bioimaging. Chem Soc Rev. 2013;42(12):5489–521. https://doi.org/10.1039/C3CS60017C.
Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. https://doi.org/10.1146/annurev.bioeng.7.060804.100432.
Chan WCW, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13(1):40–6. https://doi.org/10.1016/S0958-1669(02)00282-3.
Collins DJ, Neild A, deMello A, Liu A-Q, Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip. 2015;15(17):3439–59. https://doi.org/10.1039/C5LC00614G.
Kern KM, Schroeder JR. Comparison of cantharidin toxicity in breast cancer cells to two common chemotherapeutics. Int J Breast Cancer. 2014;2014:423059. https://doi.org/10.1155/2014/423059.
Muluneh M, Kim B, Buchsbaum G, Issadore D. Miniaturized, multiplexed readout of droplet-based microfluidic assays using time-domain modulation. Lab Chip. 2014;14(24):4638–46. https://doi.org/10.1039/C4LC00819G.
Gu S-Q, Zhang Y-X, Zhu Y, Du W-B, Yao B, Fang Q. Multifunctional picoliter droplet manipulation platform and its application in single cell analysis. Anal Chem. 2011;83(19):7570–6. https://doi.org/10.1021/ac201678g.
Bui M-PN, Li CA, Han KN, Choo J, Lee EK, Seong GH. Enzyme kinetic measurements using a droplet-based microfluidic system with a concentration gradient. Anal Chem. 2011;83(5):1603–8. https://doi.org/10.1021/ac102472a.
Acknowledgements
This work was supported by grants from the National Institute of Biomedical Imaging and Bioengineering, R03EB02935 (ATM); the National Science Foundation, CBET1509713 (ATM); and the Louisiana Board of Regents, LEQSF (2016-19)-RD-A-03 (JAD/PD). The authors would like to thank Dr. Nancy Albritton (University of North Carolina) for providing the GFP-expressing HeLa cells and Dr. Elizabeth Martin (LSU) for providing the MDA-MB-231 cells and RFP-expressing MDA-MB-231 cells. The authors would like to thank Riad Elkhanoufi (LSU) for the assistance with device fabrication.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Electronic supplementary material
ESM 1
(PDF 638 kb)
Rights and permissions
About this article
Cite this article
Vaithiyanathan, M., R. Bajgiran, K., Darapaneni, P. et al. Luminescent nanomaterials for droplet tracking in a microfluidic trapping array. Anal Bioanal Chem 411, 157–170 (2019). https://doi.org/10.1007/s00216-018-1448-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1448-1