Abstract
Key message
Iron and zinc deficiencies negatively impact human health worldwide. We developed wheat lines that meet or exceed recommended dietary target levels for iron and zinc in the grains. These lines represent useful germplasm for breeding new wheat varieties that can reduce iron and zinc deficiency-associated health burdens in the affected populations.
Abstract
Micronutrient deficiencies, including iron and zinc deficiencies, have negative impacts on human health globally. Iron-deficiency; anemia affects nearly two billion people worldwide and is the cause of reduced cognitive development, fatigue and overall low productivity. Similarly, zinc deficiency causes stunted growth, decreased immunity and increased risk of respiratory infections. Biofortification of staple crops is a sustainable and effective approach to reduce the burden of health problems associated with micronutrient deficiencies. Here, we developed wheat lines expressing rice NICOTIANAMINE SYNTHASE 2 (OsNAS2) and bean FERRITIN (PvFERRITIN) as single genes as well as in combination. NAS catalyzes the biosynthesis of nicotianamine (NA), which is a precursor of the iron chelator deoxymugeneic acid (DMA) required for long distance iron translocation. FERRITIN is important for iron storage in plants because it can store up to 4500 iron ions. We obtained significant increases of iron and zinc content in wheat grains of plants expressing either OsNAS2 or PvFERRTIN, or both genes. In particular, wheat lines expressing OsNAS2 greatly surpass the HarvestPlus recommended target level of 30 % dietary estimated average requirement (EAR) for iron, and 40 % of EAR for zinc, with lines containing 93.1 µg/g of iron and 140.6 µg/g of zinc in the grains. These wheat lines with dietary significant levels of iron and zinc represent useful germplasm for breeding new wheat varieties that can reduce micronutrient deficiencies in affected populations.
Similar content being viewed by others
Introduction
Micronutrients are essential in the human diet because they are required for key metabolic reactions and biological functions. A large fraction of the global human population suffers from micronutrient deficiencies, which has a negative impact on well-being and economic development (Muthayya et al. 2013). The most common deficiencies include iron, zinc, iodine, vitamin A, vitamin B-12, riboflavin, vitamin D, and vitamin E deficiencies (Allen et al. 2009). Iron and zinc deficiencies are among the most widespread micronutrient deficiencies. Iron deficiency is the leading cause of iron-deficiency anemia (IDA), which is prevalent world wide in both the developed and developing countries (WHO 2016). IDA affects cognitive development, decreases immune function, and causes higher mortality of mothers and children at birth. Children as well as pregnant and non-pregnant women are at a higher risk with approximately 43, 38 and 29 % of the population, respectively, affected with IDA (Stevens et al. 2013). Zinc deficiency is associated with diseases such as diarrhea, pneumonia and malaria (WHO 2002). The main clinical symptoms include growth retardation, cell-mediated immune dysfunction and cognitive impairment (Prasad 2014). Iron and zinc deficiencies are often associated with low micronutrient content in staple foods, poor micronutrient absorption from food, and dependence of the affected populations on simple and monotonous diets.
The three recommended approaches to overcome micronutrient deficiencies include supplementation, food fortification and biofortification. Micronutrient supplementation, which has been implemented in different parts of the world, is not always successful. For example, a nutritional anemia control program was implemented in India in 1970 but had only limited impact mainly because of logistical problems, poor compliance, and lack of sufficient funds (Mayer et al. 2008). This suggests that supplementation is not a sustainable preventive solution, but rather a short-term curative measure. Food fortification with iron is difficult because iron compounds such as FeSO4 cause food to have a generally unacceptable flavor and color, thus making it unpalatable (Abbaspour et al. 2014; Hurrell 2002). Biofortification of staple crops, i.e., enhancing the nutritional content of the edible parts, offers the most sustainable method to overcome micronutrient deficiencies.
Wheat (Triticum aestivum) is grown on 219 million hectares (mha) with a production of over 715 metric tonnes (MT) and is one of the most widely grown and consumed cereals globally (FAOSTAT 2013). Several billion people around the world depend on wheat for protein, starch, fiber, and various essential micronutrients (Shewry 2009). However, most of the bread wheat varieties do not contain sufficient iron and zinc in the grains to meet the daily dietary recommendations for these nutrients (Zhao et al. 2009). Therefore, biofortification of wheat for iron and zinc can greatly benefit global human health. According to the HarvestPlus program initiated by the Consultative Group for International Agricultural Research (CGIAR), 59 μg/g (dry weight) of iron and 38 μg/g (dry weight) of zinc is required in the wheat grains to meet 30 and 40 % of the estimated average requirement (EAR) of an adult diet, respectively (Bouis et al. 2011). To date, conventional breeding has achieved little or no success in enhancing the iron content of wheat. Screening of wheat germplasm showed a negative correlation between grain yield and micronutrient content (Amiri et al. 2015; Zhao et al. 2009), which might be the consequence of plant breeding focusing mainly on agronomic yield than nutritional quality in the past. Therefore, increasing grain iron and zinc content in wheat while maintaining yield has been difficult to achieve via conventional breeding. Biofortification utilizing gene technology offers a multi-dimensional approach to enrich the required micronutrients specifically in the grains without altering overall yield (Bhullar and Gruissem 2013).
Iron is important for both plants and animals. However, excessive iron accumulation is toxic to cells, and iron homeostasis is tightly regulated in animals and plants. Thus, a careful choice of strategies is required to increase iron content in cereal grains (Grusak et al. 1999). Cereals, including rice and wheat, use a chelation-based strategy (also known as Strategy II) for iron uptake (Marschner and Romheld 1994). Mugineic acid family phytosiderophores (PS) are released into the soil where they chelate the ferric form of iron, forming a PS-Fe3+ complex, which is subsequently transported into the roots by specific transporters. The PS are synthesized from S-adenosyl-l-methionine via a conserved pathway of reactions catalyzed by nicotianamine synthase (NAS), nicotianamine aminotransferase (NAAT), and deoxymugineic acid synthase (DMAS) (Kobayashi and Nishizawa 2012). Based on this knowledge various single gene and multigene strategies have been developed in rice to improve iron content in the endosperm (Bhullar and Gruissem 2013). Masuda et al. (2009) showed that constitutive overexpression of the HvNAS1 gene in rice resulted in a threefold increase of iron in the rice endosperm. Similarly, constitutive overexpression of OsNAS2 increased iron content in polished rice by 2- to fourfold (Johnson et al. 2011; Lee et al. 2012). In addition to iron, the zinc content was also increased in polished grains of NAS overexpressing lines (Johnson et al. 2011; Wirth et al. 2009). The Nicotianamine (NA) and PS can also bind Zn, in addition to Fe, thus explaining the accompanied increases in zinc content in the lines overexpressing NAS (Schaaf et al. 2004). Transformation of cereals with FERRITIN to increase the iron content has also been reported (Borg et al. 2012; Vasconcelos et al. 2003). FERRITIN is a complex of 24 protein subunits arranged to form a hollow structure that can store up to 4500 ferric molecules (Harrison and Arosio 1996). In rice, FERRITIN expressed under the control of an endosperm-specific promoter increased iron content in polished rice grains by 2- to 3.7-fold (Goto et al. 1999; Lucca et al. 2001; Oliva et al. 2014; Qu et al. 2005; Vasconcelos et al. 2003). Moreover, constitutive expression of soybean FERRITIN (GmFERRITIN) increased iron content in the leaves of wheat (Drakakaki et al. 2000). Borg et al. (2012) overexpressed endogenous wheat FERRITIN (TaFERRITIN1-A) under the control of the native endosperm-specific promoter of TaFERRITIN1-A, and reported an increase of 50–80 % of iron content in wheat grains. In rice, expression of both FERRITIN and NAS synergistically increases the grain iron content further. Endosperm-specific expression of bean PvFERRITIN and constitutive expression of Arabidopsis AtNAS1 from a single construct increased iron content in rice endosperm by sixfold (Wirth et al. 2009).
To achieve the recommended level of iron in wheat grains for healthy human diets (i.e., minimum 59 µg/g iron/dry weight; Bouis et al. 2011) it is necessary to increase iron content by 100 % in the major wheat cultivars. We have engineered bread wheat with a bean FERRITIN (PvFERRITIN) gene expressed under the control of the endosperm-specific rice GLOBULIN (OsGLOBULIN) promoter and the rice NAS2 (OsNAS2) gene expressed under the control of constitutive-promoter maize UBIQUITIN (ZmUBIQUITIN) as single gene constructs and as a combination of both genes. Expression of PvFERRITIN or OsNAS2 alone as well as in combination significantly increased iron and zinc levels in wheat grains, with several lines surpassing the recommended target levels for both iron and zinc content.
Materials and methods
Transformation vectors
PvFERRITIN (X58274) under the control of the rice endosperm-specific OsGLOBULIN promoter (Wirth et al. 2009) was excised from the parent plasmid using Xba1 and Pst1 and was used for transformation of wheat. For generation of the construct combining both PvFERRITIN and OsNAS2, we incorporated KpnI restriction site upstream of the OsGLOBULIN promoter and SmaI restriction site at 3′ of nopaline synthase (NOS) gene terminator (nosT) in the PvFERRITIN construct, and cloned it to the Pjet1.2 vector, generating PvFERRITIN-Pjet1.2 plasmid. The rice OsNAS2 (LOC_Os03g19420.2) gene was commercially synthesized along with nosT from GenScript® (http://www.genscript.com). SpeI, HindIII, and BamHI restriction sites were inserted upstream of OsNAS2, and SmaI was inserted at 3’end, and cloned into the puc57 vector, generating the OsNAS2-puc57 construct. The ZmUBIQUITIN promoter from pAHC17 (Christensen and Quail 1996) was excised using HindIII and BamHI, and was inserted into the OsNAS2-puc57 construct. The entire OsNAS2 cassette was then excised using SpeI and SmaI restriction sites, and was inserted to the PBSKII(−) vector, generating OsNAS2-PbskII(−). Similarly, the PvFERRITIN cassette was excised from the PvFERRITIN-Pjet1.2 construct using KpnI and SmaI, and cloned into the OsNAS2-PbskII(−) construct, finally generating PvFERRITIN-OsNAS2. For transformation, vector backbone-free gene cassettes were co-transformed along with the PHOSPHOMANNOSE ISOMERASE (PMI) selectable marker gene expressed under the control of the ZmUBIQUITIN promoter (Fig. 1) (Brunner et al. 2011).
Schematic illustration of the DNA constructs used for transformation. a PvFERRITIN (transformed in Fer-BW lines). b OsNAS2 (transformed in Nas-BW lines). C PvFERRITIN-OsNAS2 (transformed in FerNas-BW lines). D ZmUBIQUITIN-PMI (co-transformed with constructs a, b and c). Numbers indicate length of the gene cassette in base pairs. For details see “Materials and methods”
Wheat transformation, plant growth and characterization
Hexaploid spring wheat cultivar Bobwhite SH 98 26 was transformed using particle gun bombardment (Brunner et al. 2011; Pellegrineschi et al. 2002). Overall, approximately 6500 immature embryos were co-transformed with PMI and PvFERRITIN, PMI and OsNAS2, or PMI and PvFERRITIN-OsNAS2 generating transformed lines named PvFERRITIN-Bobwhite (hereafter Fer-BW), OsNAS2-Bobwhite (hereafter Nas-BW), and PvFERRITIN-OsNAS2-Bobwhite (hereafter FerNas-BW), respectively. Selection of the transformed plants was performed on culture media containing mannose during the regeneration phase (Wright et al. 2001). Plants were later grown in commercial soil (Klasmann-Deilmann GmbH, Germany) in the greenhouse in 16 h light/22 °C and 8 h dark/18 °C, and a humidity of 60 %. Genomic DNA was isolated from 3-week-old seedlings in the T0-, T1-, T2- and T3-generation (Stein et al. 2001; Vasudevan et al. 2014). PCR was used to confirm the presence of the transgenes using gene specific primers (Table. S1). PCR-positive lines were further selected based on the iron content in the subsequent generations. Southern blot analysis was performed using a P32-labelled probe to select lines with a single copy insert (Fig. S1) in T0-generation plants (Green and Sambrook 2012). The probes for PvFERRITIN, OsNAS2, and PMI were generated using gene specific primers (Table. S1). For phenotypic characterization of transformed wheat lines, parameters including days to flowering (DTF), plant height, 1000 grain weight (1000 GW), and tiller number were recorded.
Metal ion measurements
Plants were harvested 6 weeks after flowering and spikes were dried at 37 °C for 3 days. Grain samples were de-husked and grounded for metal ion measurements. Additionally, ground grain samples were fractioned using a 250 μm nylon sieve to obtain sieved flour (Borg et al. 2012), referred to as ‘flour’ hereafter. Two hundred mg of sample was boiled in 15 ml of 65 % v/v HNO3 solution at 120 °C for 90 min. Three ml of 30 % v/v H2O2 was added and boiled at 120 °C for 90 min. Metal concentrations were determined using inductively coupled plasma-optical emission spectroscopy (ICP-OES) (Varian Vista-MPX CCD Simultaneous ICP-OES). The wavelength used for iron, zinc, copper, manganese, and magnesium was 238.204, 213.857, 324.754, 257.610, and 285.213, respectively. The iron concentrations were recorded in T2, T3 and T4 grains, and the seeds from plants with highest iron concentration were used to grow the next generation of plants (Fig. S2). Data were analyzed using the Student’s t test to determine statistically significant differences among the transformed lines and their respective controls.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed to assess the expression of the transgenes in the transgenic lines. Total RNA was extracted from leaves and grains collected 18 days after flowering (DAF) in the T3 generation plants. Leaf RNA was extracted using Trizol® reagent (Invitrogen, USA), and the RNA was treated with DNase I (Thermo Fisher Scientific, Inc., USA) to remove genomic DNA contamination. cDNA synthesis was done using the RevertAid™ first strand cDNA synthesis kit (Thermo Fisher Scientific, Inc., USA). qRT-PCR was performed on the 7500 FAST Real Time PCR system (Applied Biosystems, Inc., USA). The total reaction volume of 20 µl included 1 µl cDNA, 0.4 µl forward primer, 0.4 µl reverse primer, 10 µl Sybrgreen Mastermix (Applied Biosystems, Ltd., USA), and 8.2 µl H2O. Primers were designed using a CLC Genomics Workbench (Table S1). For data normalization, Ta.22845 was used as reference gene that encodes for ATP-dependent 26S proteasome regulatory subunit (Paolacci et al. 2009), and it expresses both in the flag leaves as well as the grains of wheat (own unpublished data). The Ct value was obtained from 7500 Fast System Software. The data normalization was done as described by Liu et al. (2011).
Results
Seed specific expression of PvFERRITIN and constitutive expression of OsNAS2 in wheat
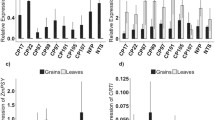
Single insertion Fer-BW, Nas-BW, and FerNas-BW transgenic lines (Figs. 1, S1) were analyzed for transgene expression in grains and leaves. PvFERRITIN was specifically expressed in the grains of all Fer-BW and FerNas-BW lines, except for two lines, Fer-BW 51 and FerNas-BW 30, which also showed expression in leaves (Figs. 2a, c, S3). These results confirm the seed-specific expression of PvFERRITIN under the control of the rice OsGLOBULIN promoter in most wheat lines. OsNAS2-specific primers (Table S1) were used to analyze the expression of the transgene in Nas-BW leaves and both leaves and grains of FerNas-BW lines (Fig. 2b, d). All Nas-BW lines showed OsNAS2 expression in the leaves. In FerNas-BW lines OsNAS2 expression was also detected in grains, except for FerNas-BW 17. However, OsNAS2 expression was many folds higher in leaves than grains, indicating that the ZmUBIQUITIN promoter is not very active in wheat endosperm.
Relative expression of transgenes PvFERRITIN and OsNAS2 at 18 days after flowering (DAF) in T3-generations. a Relative expression of PvFERRITIN in grains and leaves of Fer-BW lines. b Relative expression of OsNAS2 in leaves of Nas-BW lines. c Relative expression of PvFERRITIN in FerNas-BW lines. d Relative expression of OsNAS2 in FerNas-BW lines. No expression of PvFERRITIN and OsNAS2 was observed in Bobwhite (BW) and non-transgenic sibling (NTS) controls. The data were normalized to the endogenous expression of Ta.22845. Values are the average of three biological replicates (±SD)
Expression of PvFERRITIN increases iron and zinc content in whole grains and flour
The single insertion Fer-BW lines 18, 47, 51, 62, and 69 were analyzed for their metal content. Iron content was significantly increased in all lines except Fer-BW 69. Iron content in the T4 grains of these lines ranged between 42.0 and 61.9 µg/g DW, as compared to 38.5 µg/g DW in Bobwhite (Fig. 3). Fer-BW 51 had the highest iron content (61.9 µg/g) with an increase of 1.6-fold as compared to the control grains. The iron content in the flour (the endosperm of the grains) of Fer-BW transgenic lines ranged from 20.2 to 33.9 µg/g DW compared to 20.4 µg/g DW in Bobwhite. Flour from grains of line 47 had the highest iron content, with a 1.7-fold increase (33.6 µg/g) as compared to the control (Fig. 3). In addition, whole grains of lines 18, 47, 51 and 62 had significantly increased zinc content ranging from 59.1 to 72.6 µg/g DW as compared to 43.1 µg/g DW in Bobwhite (Figs. 3, S4). Zinc content in the flour ranged from 30.7 to 35.3 µg/g DW in the transformed lines as compared to 23.0 µg/g DW in the control. Line Fer-BW 47, which contained the highest iron content in the flour, also contained the highest zinc content in whole grains (72.6 µg/g) as well as in flour (35.3 µg/g), representing 1.7-fold and 1.5-fold increases, respectively, as compared to the control. In addition, copper, manganese, and magnesium levels were also measured in whole grains and flour. Copper was increased in grains of four of the Fer-BW lines, except for line 69 (Fig. S5), and ranged from 6.9 to 8.3 µg/g DW compared to 6.5 µg/g DW in the control line. A similar trend was observed in the copper content of flour which ranged from 4.8 to 5.7 µg/g DW in the transformed lines, compared to 3.91 µg/g DW in Bobwhite. With few exceptions, manganese and magnesium content was not altered in the transformed lines (Fig. S5). Phenotypic greenhouse performance of the Fer-BW lines were similar to Bobwhite for plant height and 1000 GW, while they showed some variation for days to flowering (DTF) and tiller number (Table. S2).
Iron (Fe) and zinc (Zn) content in T4 grains and flour of lines expressing PvFERRITIN (Fer-BW). Values are the mean of three biological replicates (±SD). Black asterisks above the bars indicate statistically significant values calculated using Student’s T test, in comparison to the control line Bobwhite (BW) (*P < 0.05, **P < 0.01). NTS non-transgenic sibling
OsNAS2 overexpression increases iron and zinc content to dietary significant levels
Ten single insertion Nas-BW lines expressing rice OsNAS2 had significant increases in iron and zinc content in comparison to Bobwhite (Fig. 4). The iron content in T4 whole grains ranged from 59.6 to 93.1 µg/g DW, as compared to 42.7 µg/g DW in the control. Moreover, iron content in the flour of these lines ranged from 30.1 to 53.3 µg/g DW, as compared to 21.4 µg/g DW in the control. Line 65 had the greatest increase in iron (2.1-fold) in whole grains and a 2.5-fold increase in the flour as compared to Bobwhite. The Nas-BW lines had relatively higher iron content in comparison to Fer-BW lines and all of these lines also surpass the suggested target levels of 59 µg/g DW iron in whole grains to meet the 30 % estimated average requirement of human diets. The iron increases in the NAS-BW lines correlated well with the transgene expression (Fig. 2) in these plants, with line 65 having the highest OsNAS2 expression in leaves.
Iron (Fe) and zinc (Zn) content in T4 grains and flour of lines expressing OsNAS2 (Nas-BW). Values are the mean of three biological replicates (± SD). Black asterisks above the bars indicate statistically significant values calculated using Student’s T test, in comparison to the control line Bobwhite (BW) (*P < 0.05; **P < 0.01). NTS non-transgenic sibling
The zinc content in whole grains of the transformed lines ranged from 54.2 to 140.6 µg/g DW and in the flour from 31.9 to 94.1 µg/g DW, as compared to 37.9 and 22.4 µg/g DW in Bobwhite, respectively (Fig. 4). Line 40 had the greatest zinc levels with a 3.7-fold increase (140.6 µg/g) in whole grains and a 4.2-fold increase (94.1 µg/g) in flour, as compared to control. Increases in copper, magnesium, and manganese content were also measured in most of the transformed lines (Fig. S6). All lines had significantly higher copper content in whole grains as well as flour. The manganese content also increased significantly in whole grains and flour in most of the lines except in whole grains of lines 38, 53, 66 and 81 and flour of line 66. As an exception, line 91 showed significantly lower manganese content in both whole grains and flour. The magnesium content in whole grains as well as flour was significant increased except in whole grains of lines 42 and 66 and flour of line 91.
Importantly, most of the Nas-BW lines were phenotypically indistinguishable from Bobwhite for DTF, plant height, 1000 GW, and tiller number (Table 1). Lines 40 and 91 flowered earlier, and 1000 GW of lines 40 and 42 was significantly increased but decreased in line 81 as compared with Bobwhite.
Iron and zinc levels in lines expressing both PvFERRITIN and OsNAS2
Wheat lines expressing PvFERRITIN and OsNAS2 from a single construct (FerNas-BW lines) were analyzed to determine if the transgenes act synergistically leading to higher iron and zinc increases in comparison to plants expressing single genes (Fer-BW and Nas-BW lines). The iron content in whole grains of five single insertion FerNas-BW lines ranged from 35.6 to 60.3 µg/g DW, as compared to 42.7 µg/g DW in Bobwhite (Fig. 5). The flour of these transgenic lines had iron content in the range of 24.9–45.3 µg/g DW, compared to 21.4 µg/g DW in Bobwhite. FerNas-BW lines had a maximum 1.7-fold increased of iron (60.3 µg/g in line 15) in whole grains and of 1.8-fold increased iron in the flour (45.3 µg/g in line 3), as compared to Bobwhite. Among the FerNas-BW lines, line 3 had the highest zinc content in whole grains (82.4 µg/g DW) as well as in flour (55.7 µg/g DW) (Fig. 5). However, in contrary to the expectations, these increases for iron and zinc content are smaller than those observed in plants expressing either PvFERRITIN or OsNAS2 alone. These lower increases in iron and zinc content possibly correlate to a relatively lower level of FERRITIN expression in the FerNas-BW lines as compared to that in the Fer-BW lines.
Iron (Fe) and zinc (Zn) content in T4 grains and flour of lines expressing PvFERRITIN-OsNAS2 (FerNas-BW). Values are the mean of three biological replicates (±SD). Black and red asterisks above the bars indicate statistically higher and lower significant values calculated using Student’s T test, respectively, in comparison to the control line Bobwhite (BW) (*P < 0.05; **P < 0.01). NTS non-transgenic sibling
Copper, manganese and magnesium contents were variable among the different FerNas-BW lines (Fig. S7). Lines 3, 15, and 42 showed significantly increased copper and magnesium content in both whole grains as well as the flour. In contrast, lines 17 and 30 had reduced copper content in comparison to the control. Manganese content was also significantly higher in lines 3 and 15 for both the whole grains and the flour, as compared with Bobwhite. FerNas-BW lines were phenotypically similar to Bobwhite for DTF, 1000 GW and tiller number as well as plant height, except for line 42 which was significantly taller (Table S3).
Discussion
The development of high yielding farmer-preferred wheat cultivars with increased micronutrient content in the grains can improve the well-being of millions of malnourished humans around the globe. While increases for some micronutrients could be achieved via conventional breeding, the improvements for many other micronutrients including iron, vitamin A and folate are difficult to achieve (Bhullar and Gruissem 2013; Mayer et al. 2008; Slamet-Loedin et al. 2015). In 2015, zinc biofortified wheat varieties were released in India and Pakistan (HarvestPlus 2015a, b). Zinc fertilizer application has also been suggested as a useful strategy for zinc biofortification of wheat (Cakmak 2008). Similar agronomic practices as well as conventional breeding approaches, however, have not been successful in achieving the recommended increase of iron content in wheat grains. Furthermore, it is difficult to implement targeted fertilizer application strategies, particularly in the developing countries, because it is impractical and expensive. Thus, engineering of wheat lines with high-iron and -zinc grains is a more economical and sustainable solution for micronutrient improvement and human health. As we show here, iron biofortification of wheat using gene technology is a robust method for approaching or achieving the required target levels. So far, only few genetic engineering attempts have been made to biofortify wheat for increased iron content in the grains (Drakakaki et al. 2000; Borg et al. 2012; Sui et al. 2012). We focused on two different strategies using constitutive overexpression of NAS and endosperm-specific expression of FERRITIN either alone or combined. The results we obtained for the Fer-BW lines expressing PvFERRITIN (Fig. 3) are similar to those previously reported (Borg et al. 2012; Sui et al. 2012). The increased zinc content in the Fer-BW lines (Fig. 3) is consistent with previous reports of expressing FERRITIN in both wheat and rice (Borg et al. 2012; Trijatmiko et al. 2016; Vasconcelos et al. 2003; Wirth et al. 2009).
In contrast to the Fer-BW lines, the lines overexpressing OsNAS2 had higher iron and zinc increases in the grains, with selected lines surpassing the suggested iron target of 59 µg/g to meet the recommended 30 % EAR (Fig. 4). These lines also have higher zinc contents. Most importantly, the high whole grain iron and zinc increases are retained in the flour (Fig. 4), which usually is the part of the grain that is consumed. Overexpression of NAS increases the production of nicotianamine (NA) and deoxymugineic acid (DMA), ultimately facilitating the uptake and transport of PS-Fe3+ chelates (Wang et al. 2013; Wirth et al. 2009). NAS overexpression in rice also increases the expression of genes encoding the zinc transporters OsZIP1 and OsZIP4 (Wang et al. 2013), which can explain the increased zinc content of the transgenic lines. NA is also involved in long-distance iron transport in the phloem and translocation to the grains (Puig et al. 2007). Constitutive expression of OsNAS2 also increased iron content in polished rice (Johnson et al. 2011; Lee et al. 2012). Moreover, mugineic acid (MA) and NA function in the uptake of other essential micronutrients, including zinc, copper, and manganese (Haydon and Cobbett 2007). Collectively, the significant increases in copper and manganese content that we find in nearly all of the Nas-BW lines in whole grains and as well as flour is consistent with the reported data (Fig. S6).
Unexpectedly, however, the expression of both PvFERRITIN and OsNAS2 in wheat does not result in higher iron levels when compared to lines expressing OsNAS2 alone. Thus, unlike rice in which the expression of FERRITIN and NAS increased iron content by sixfold in the polished rice grains (Trijatmiko et al. 2016; Wirth et al. 2009), the two genes do not seem to function synergistically in wheat. Although the reasons for this are unclear, it is plausible that iron homeostasis is controlled differently in hexaploid wheat as compared to rice and that iron accumulation is tightly regulated. Furthermore, we did not find a correlation between PvFERRITIN expression and iron content in both Fer-BW and FerNas-BW transgenic lines (Fig. 2) while OsNAS2 expression in leaves correlated with the iron content of Nas-BW and FerNas-BW lines.
Together, we found that constitutive expression of OsNAS2 alone is the preferred strategy for nutritionally relevant iron biofortification in wheat. Our data suggest that unsuccessful breeding efforts to increase iron in wheat grains could be the consequence of regulated iron uptake and transport, which can be overcome by constitutive expression of OsNAS2. Further detailed molecular analysis of the transgenic lines is required to understand the mechanism of metal homeostasis in wheat and how this is changed in the transgenic lines to increase grain iron content. Understanding the consequences of constitutive OsNAS2 expression on endogenous wheat metal homeostasis-related genes will help to expand our knowledge of their interactions and will assist in improving iron in other preferred crops as well. The high iron and zinc wheat lines that we have developed are a promising breeding material after further agronomic performance testing to confirm that the iron and zinc increases observed in the greenhouse conditions are maintained in the field. The high iron and zinc traits could be easily bred into farmer-preferred and commercial cultivars, thereby reducing iron and zinc micronutrient deficiencies in affected populations.
Author contribution statement
N. K. B. conceived and designed the experiments. S.S. carried out the experiments. S. S. and N. K. B. analyzed the data. S.S., N.K.B., B.K and W.G. discussed the data. S. S. and N. K. B. wrote the manuscript. W. G. and N. K. B. edited the manuscript. All authors have read the manuscript and agree with its content.
References
Abbaspour N, Hurrell R, Kelishadi R (2014) Review on iron and its importance for human health. J Res Med Sci 19:164–174
Allen LH, Peerson JM, Olney DK (2009) Provision of multiple rather than two or fewer micronutrients more effectively improves growth and other outcomes in micronutrient-deficient children and adults. J Nutr 139:1022–1030. doi:10.3945/jn.107.086199
Amiri R, Bahraminejad S, Sasani S, Jalali-Honarmand S, Fakhri R (2015) Bread wheat genetic variation for grain’s protein, iron and zinc concentrations as uptake by their genetic ability. Eur J Agron 67:20–26. doi:10.1016/j.eja.2015.03.004
Bhullar NK, Gruissem W (2013) Nutritional enhancement of rice for human health: the contribution of biotechnology. Biotechnol Adv 31:50–57. doi:10.1016/j.biotechadv.2012.02.001
Borg S, Brinch-Pedersen H, Tauris B, Madsen LH, Darbani B, Noeparvar S, Holm PB (2012) Wheat ferritins: improving the iron content of the wheat grain. J Cereal Sci 56:204–213. doi:10.1016/J.Jcs.2012.03.005
Bouis HE, Hotz C, McClafferty B, Meenakshi JV, Pfeiffer WH (2011) Biofortification: a new tool to reduce micronutrient malnutrition. Food Nutr Bull 32:S31–S40
Brunner S et al (2011) Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol J 9:897–910. doi:10.1111/j.1467-7652.2011.00603.x
Cakmak I (2008) Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 302:1–17. doi:10.1007/s11104-007-9466-3
Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218. doi:10.1007/Bf01969712
Drakakaki G, Christou P, Stoger E (2000) Constitutive expression of soybean ferritin cDNA intransgenic wheat and rice results in increased iron levels in vegetative tissues but not in seeds. Transgenic Res 9:445–452. doi:10.1023/A:1026534009483
FAOSTAT (2013). http://faostat3.fao.org/. Accessed 7 October 2015
Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17:282–286
Green MR, Sambrook J (2012) Molecular cloning a laboratory manual, 4th edn. Cold Spring Harbor Laboratory Press, New York
Grusak MA, Pearson JN, Marentes E (1999) The physiology of micronutrient homeostasis in field crops Field Crop Res 60:41–56. doi:10.1016/S0378-4290(98)00132-4
Harrison PM, Arosio P (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275:161–203
HarvestPlus (2015a) India Country brief. http://www.harvestplus.org/sites/default/files/HarvestPlus_CountryBrief_India_0.pdf. Accessed 6 April 2016
HarvestPlus (2015b) Pakistan Country brief. http://www.harvestplus.org/sites/default/files/HarvestPlus_CountryBrief_Pakistan_1.pdf
Haydon MJ, Cobbett CS (2007) Transporters of ligands for essential metal ions in plants. New Phytol 174:499–506. doi:10.1111/j.1469-8137.2007.02051.x
Hurrell R (2002) How to ensure adequate iron absorption from iron-fortified food. Nutr Rev 60:S7–S15
Johnson AAT, Kyriacou B, Callahan DL, Carruthers L, Stangoulis J, Lombi E, Tester M (2011) Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS One. doi:10.1371/journal.pone.0024476
Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Ann Rev Plant Biol 63:131–152
Lee S, Kim YS, Jeon US, Kim YK, Schjoerring JK, An G (2012) Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol Cells 33:269–275. doi:10.1007/S10059-012-2231-3
Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1 s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34:738–751. doi:10.1111/j.1365-3040.2011.02278.x
Lucca P, Ye XD, Potrykus I (2001) Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol Breed 7:43–49. doi:10.1023/A:1009661014167
Marschner H, Romheld V (1994) Strategies of plants for acquisition of iron. Plant Soil 165:261–274. doi:10.1007/Bf00008069
Masuda H et al (2009) Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2:155–166. doi:10.1007/s12284-009-9031-1
Mayer JE, Pfeiffer WH, Beyer P (2008) Biofortified crops to alleviate micronutrient malnutrition. Curr Opin Plant Biol 11:166–170. doi:10.1016/j.pbi.2008.01.007
Muthayya S, Rah JH, Sugimoto JD, Roos FF, Kraemer K, Black RE (2013) The global hidden hunger indices and maps: an advocacy tool for action. PLoS One. doi:10.1371/journal.pone.0067860
Oliva N et al (2014) Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol Breed 33:23–37. doi:10.1007/s11032-013-9931-z
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11. doi:10.1186/1471-2199-10-11
Pellegrineschi A et al (2002) Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 45:421–430. doi:10.1139/G01-154
Prasad AS (2014) Impact of the discovery of human zinc deficiency on health. J Trace Elem Med Biol 28:357–363. doi:10.1016/j.jtemb.2014.09.002
Puig S, Andres-Colas N, Garcia-Molina A, Penarrubia L (2007) Copper and iron homeostasis in arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30:271–290. doi:10.1111/j.1365-3040.2007.01642.x
Qu LQ, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222:225–233. doi:10.1007/S00425-005-1530-8
Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279:9091–9096. doi:10.1074/jbc.M311799200
Shewry PR (2009) Wheat J Exp Bot 60:1537–1553. doi:10.1093/jxb/erp058
Slamet-Loedin IH, Johnson-Beebout SE, Impa S, Tsakirpalogloul N (2015) Enriching rice with Zn and Fe while minimizing Cd risk Front. Plant Sci. doi:10.3389/fpls.2015.00121
Stein N, Herren G, Keller B (2001) A new DNA extraction method for high-throughput marker analysis in a large-genome species such as Triticum aestivum. Plant Breed 120:354–356. doi:10.1046/j.1439-0523.2001.00615.x
Stevens GA et al (2013) Global, regional, and national trends in haemoglobin concentration and prevalence of total and severe anaemia in children and pregnant and non-pregnant women for 1995-2011: a systematic analysis of population-representative data. Lancet Glob Health 1:e16–e25. doi:10.1016/S2214-109X(13)70001-9
Sui X, Zhao Y, Wang S, Duan X, Xu L, Liang R (2012) Improvement Fe content of wheat(Triticum aestivum) grain by soybean ferritin expression cassette without vector backbone sequence J Agric. Biotechnol 20:766–773
Trijatmiko KR et al (2016) Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci Rep Uk. doi:10.1038/srep19792
Vasconcelos M et al (2003) Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci 164:371–378. doi:10.1016/S0168-9452(02)00421-1
Vasudevan K, Cruz CMV, Gruissem W, Bhullar NK (2014) Large scale germplasm screening for identification of novel rice blast resistance sources Front. Plant Sci. doi:10.3389/fpls.2014.00505
Wang M, Gruissem W, Bhullar NK (2013) Nicotianamine synthase overexpression positively modulates iron homeostasis-related genes in high iron rice Front. Plant Sci. doi:10.3389/fpls.2013.00156
WHO (2002) The world health report 2002—reducing risks, promoting healthy life. Switzerland, Geneva
WHO (2016) Micronutrient deficiency: iron deficiency anaemia. http://www.who.int/nutrition/topics/ida/en/. Accessed 06 Feb 2016
Wirth J et al (2009) Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol J 7:631–644. doi:10.1111/J.1467-7652.2009.00430.X
Wright M et al (2001) Efficient biolistic transformation of maize (Zea mays L.) and wheat (Triticum aestivum L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep 20:429–436
Zhao FJ, Su YH, Dunham SJ, Rakszegi M, Bedo Z, McGrath SP, Shewry PR (2009) Variation in mineral micronutrient concentrations in grain of wheat lines of diverse origin. J Cereal Sci 49:290–295. doi:10.1016/j.jcs.2008.11.007
Acknowledgments
This research was supported by ETH Zurich to W. G. and N. K. B., and from the State Secretariat for Education, Research, and Innovation (SERI) to N. K. B. We thank Gabriele Buchmann and Gerhard Herren for their support in performing the wheat transformation. We acknowledge Prof. Rainer Schulin (Soil Protection Group, ETH Zurich) for extending access to ICP-OES, and Björn Studer for guidance while performing the metal measurements. We also acknowledge the help of Irene Zurkirchen and Ting-Ying Wu for technical support in the greenhouse and the laboratory. We thank Dr. Christof Sautter for providing the PvFERRITIN expressing construct.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical statement
This work does not contain any study with human participants and/or animals, and fully complies with ethical standards.
Additional information
Communicated by S. Dreisigacker.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Singh, S.P., Keller, B., Gruissem, W. et al. Rice NICOTIANAMINE SYNTHASE 2 expression improves dietary iron and zinc levels in wheat. Theor Appl Genet 130, 283–292 (2017). https://doi.org/10.1007/s00122-016-2808-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2808-x