Abstract
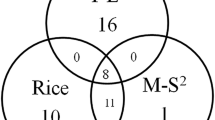
The use of plant disease resistance (R) genes in breeding programs needs an understanding of their variation patterns. In our current study, we investigated the polymorphisms of 44 NBS-LRR class R-genes among 21 rice cultivars and 14 wild rice populations. Our data suggested that there were four basic types of variations: conserved, diversified, intermediate-diversified, and present/absent patterns. Common characteristics at a locus of conserved R-genes were: copy-number uniformity, clear divergence (long branches) with other paralogs, and highly identical alleles. On the other hand, copy-number variability, a nearly equal and non-zero branch lengths, and high levels of nucleotide diversity were observed at the loci of highly diversified R-genes. Research suggests that the ratio of diverse alleles to the total number of genes at a locus is one of the best criteria to characterize the variation pattern of an R-gene. Our data suggested that a significant genetic reduction was detected only in four present/absent R-genes, compared with the variation observed in wild rice. In general, no difference was detected between wild rice and cultivars, japonica and indica rice, or between lines from different geographic regions. Our results also suggested that R-genes were under strong selection, which shaped R-gene variation patterns.



Similar content being viewed by others
References
Allen RL, Bittner-Eddy P, Grenville-Briggs L, Meitz J, Rehmany AP, Rose LE, Beynon JL (2004) Host–parasite coevolutionary conflict between Arabidopsis and Downy Mildew. Science 306:1957–1960
Bakker EG, Toomajian C, Keritman M, Bergelson J (2006) A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18:1803–1818
Bergelson J, Kreitman M, Stah EA, Tian D (2001) Evolutionary dynamics of plant R-genes. Science 292:2281–2285
Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, Mcadams SA, Faulk KN, Donaldson K, Tarchini T, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant cell 12:2033–2046
Delseny M, Salses J, Cooke R, Sallaud C, Regad F et al (2001) Rice genomics: present and future. Plant Physiol Biochem 39:323–334
Ding J, Cheng H, Jin X, Araki H, Yang Y, Tian D (2007a) Contrasting patterns of evolution between allelic groups at a single locus in Arabidopsis. Genetica 129:235–242
Ding J, Zhang W, Jing Z, Chen JQ, Tian D (2007b) Unique pattern of R-gene variation within populations in Arabidopsis. Mol Genet Genomics 277:619–629
Dodds PN, Lawrence GJ, Catanzariti A, Ayliffe MA, Ellis JG (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16:755–768
Elli J, Dodds P, Pryor T (2000) Structure function and evolution of plant disease resistance genes. Curr Opin Plant Biol 3:278–284
Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506
Garris A, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Hammond-Kosack KE, Jones JD (1997) Plant disease resistance genes Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Innan H, Kim Y (2004) Pattern of polymorphism after strong artificial selection in a domestication event. Proc Natl Acad Sci USA 101:10667–10672
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jia Y, Bryan GT, Farrall L, Valent B (2003) Natural variation at the Pi-ta rice blast resistance locus. Phytopathology 93:1452–1459
Jiang H, Wang C, Ping L, Tian D, Yang S (2007) Pattern of LRR nucleotide variation in plant resistance genes. Plant Sci 173:253–261
Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defenses. Adv Bot Res 24:89–167
Kuang H, Woo SS, Meyers BC, Nevo E, Michelmor RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16:2870–2894
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
McDowell JM, Woffenden BJ (2003) Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol 21:178–182
McNally KL, Bruskiewich R, Mackill D, Buell CR, Leach JE, Leung H (2006) Sequencing multiple and diverse rice varieties connecting whole-genome variation with phenotypes. Plant Physiol 141:26–31
Meyers BC, Kozik A , Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Maynard-Smith J, Haigh J (1974) The hitch-hiking effect of a favorable gene. Genet Res 23:23–35
Nasu S, Suzuki J, Ohta R, Hasegawa K, Yui R, Kitazawa N, Monna L, Minobe Y (2002) Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP Markers. DNA Res 9:163–171
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Noël L, Moores TL, Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JDG (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11:2099–2111
Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91:821–832
Pink DAC (2002) Strategies using genes for non-durable resistance. Euphytica 124:227–236
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL (2006) Nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1194
Rozas J, Sánchez-delBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Shen J, Araki H, Chen L, Chen JQ, Tian D (2006) Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172:1243–1250
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang GL, Ruan DL, Song WY, Sideris S, Chen L, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut BS, Whalen MC, Ronald PC (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10:756–779
Wang X, Shi X, Hao B, Ge S, Luo J (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165:937–946
Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19:55–64
Wichmann G, Ritchie D, Kousik CS, Bergelson J (2005) Reduced genetic variation occurs among genes of the highly clonal plant pathogen Xanthomonas axonopodis pv. vesicatoria, including the effector gene avrBs2. Appl Environ Microbiol. 71:2418–2432
Yang S, Feng Z, Zhang X, Jiang K, Jin X, Hang Y, Chen JQ, Tian D (2006) Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62:181–193
Yang S, Jiang K, Araki H, Ding J, Yang YH, Tian D (2007) A molecular isolation mechanism associated with high intra-specific diversity in rice. Gene 394:87–95
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in rice reveals significant expansion of divergent non-TIR NBS Genes. Mol Genet Genomics 271:402–415
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30570987 and 30470122), pre-program for NBRPC (2005CCA02100), 111 project and SUR grant from IBM to D.T., Y.H. or J.Q.C.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by D. A. Lightfoot.
Sihai Yang and Tingting Gu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, S., Gu, T., Pan, C. et al. Genetic variation of NBS-LRR class resistance genes in rice lines. Theor Appl Genet 116, 165–177 (2008). https://doi.org/10.1007/s00122-007-0656-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0656-4