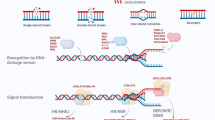
Abstract
DNA damage response (DDR) relies on swift and accurate signaling to rapidly identify DNA lesions and initiate repair. A critical DDR signaling and regulatory molecule is the posttranslational modification poly(ADP-ribose) (PAR). PAR is synthesized by a family of structurally and functionally diverse proteins called poly(ADP-ribose) polymerases (PARPs). Although PARPs share a conserved catalytic domain, unique regulatory domains of individual family members endow PARPs with unique properties and cellular functions. Family members PARP-1, PARP-2, and PARP-3 (DDR–PARPs) are catalytically activated in the presence of damaged DNA and act as damage sensors. Family members tankyrase-1 and closely related tankyrase-2 possess SAM and ankyrin repeat domains that regulate their diverse cellular functions. Recent studies have shown that the tankyrases share some overlapping functions with the DDR–PARPs, and even perform novel functions that help preserve genomic integrity. In this review, we briefly touch on DDR–PARP functions, and focus on the emerging roles of tankyrases in genome maintenance. Preservation of genomic integrity thus appears to be a common function of several PARP family members, depicting PAR as a multifaceted guardian of the genome.
Similar content being viewed by others
References
Hoeijmakers JHJ (2009) DNA damage, aging, and cancer. N Engl J Med 361:1475–1485
Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204
Wei H, Yu X (2016) Functions of PARylation in DNA damage repair pathways. Genom Proteom Bioinform 14:131–139
Aredia F, Scovassi AI (2014) Poly(ADP-ribose): a signaling molecule in different paradigms of cell death. Biochem Pharmacol 92:157–163
Schuhwerk H, Atteya R, Siniuk K, Wang ZQ (2017) PARPing for balance in the homeostasis of poly(ADP-ribosyl)ation. Semin Cell Dev Biol 63:81–91
Krishnakumar R, Kraus WL (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39:8–24
Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F (2014) Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res 329:18–25
Ray Chaudhuri A, Nussenzweig A (2017) The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 18:610–621
Farmer H et al (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917–921
Bryant HE et al (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913–917
Smith S, Giriat I, Schmitt A, de Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484–1487
Huang S et al (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620
Smith S, De Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10:1299–1302
Chambon P, Weill JD, Mandel P (1963) Nicotinamide mononucleotide activation of a new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun 11:39–43
Ruf A, Rolli V, de Murcia G, Schulz GE (1998) The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol 278:57–65
Crawford K, Bonfiglio JJ, Mikoč A, Matic I, Ahel I (2018) Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit Rev Biochem Mol Biol 53:64–82
de Murcia JM et al (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 22:2255–2263
Alano CC et al (2010) NAD + depletion is necessary and sufficient for PARP-1—mediated neuronal death. J Neurosci 30:2967–2978
Yu S-W et al (2006) Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA 103:18314–18319
Vyas S et al (2015) Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun 5:4426
Marsischky GT, Wilson BA, Collier RJ (1995) Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation: evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem 270:3247–3254
Kleine H et al (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32:57–69
Rippmann JF, Damm K, Schnapp A (2002) Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J Mol Biol 323:217–224
Dawicki-McKenna JM et al (2015) PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol Cell 60:755–768
Langelier MF, Pascal JM (2013) PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr Opin Struct Biol 23:134–143
Langelier MF, Zandarashvili L, Aguiar PM, Black BE, Pascal JM (2018) NAD + analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun 9:844
Langelier MF, Planck JL, Roy S, Pascal JM (2012) Structural basis for DNA damage—dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336:728–732
Langelier MF, Riccio A, Pascal JM (2014) PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res 42:7762–7775
Riccio AA, Cingolani G, Pascal JM (2015) PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res 44:1691–1702
Caldecott KW (2014) DNA single-strand break repair. Exp Cell Res 329:2–8
Chang HHY, Pannunzio NR, Adachi N, Lieber MR (2017) Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18:495–506
Gagné JP et al (2008) Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36:6959–6976
Ariumi Y et al (1999) Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene 18:4616–4625
Boulton SJ (2002) Combined functional genomic maps of the C. elegans DNA damage response. Science 295:127–131
Li M, Lu LY, Yang CY, Wang S, Yu X (2013) The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev 27:1752–1768
Hochegger H et al (2006) Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J 25:1305–1314
Sugimura K, Takebayashi SI, Taguchi H, Takeda S, Okumura K (2008) PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol 183:1203–1212
Ying S, Hamdy FC, Helleday T (2012) Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res 72:2814–2821
Illuzzi G et al (2014) PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucleic Acids Res 42:7776–7792
Yang Y-G, Cortes U, Patnaik S, Jasin M, Wang Z-Q (2004) Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23:3872–3882
Audebert M, Salles B, Calsou P (2004) Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279:55117–55126
Wang M et al (2006) PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34:6170–6182
Ahel D et al (2009) Poly(ADP-ribose)–dependent regulation of DNA repair by the chromatin remodeling Enzyme ALC1. Science 325:1240–1243
Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM (2010) The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem 285:18877–18887
Bryant HE et al (2009) PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28:2601–2615
Haince JF et al (2008) PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 283:1197–1208
Ko HL, Ren EC (2012) Functional aspects of PARP1 in DNA repair and transcription. Biomolecules 2:524–548
Jubin T et al (2017) Poly ADP-ribose polymerase-1: beyond transcription and towards differentiation. Semin Cell Dev Biol 63:167–179
Kraus WL, Lis JT (2003) PARP goes transcription. Cell 113:677–683
Chi NW, Lodish HF (2000) Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem 275:38437–38444
Bae J, Donigian JR, Hsueh AJW (2003) Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem 278:5195–5204
De Rycker M, Venkatesan RN, Wei C, Price CM (2003) Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem J 372:87–96
Guettler S et al (2011) Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell 147:1340–1354
Chang P, Coughlin M, Mitchison TJ (2005) Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol 7:1133–1139
Smith S, de Lange T (1999) Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci 112:3649–3656
Leung AKL et al (2011) Article poly (ADP-Ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42:489–499
Lyons RJ et al (2001) Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J Biol Chem 276:17172–17180
Chiang YJ et al (2008) Tankyrase 1 and Tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One 3:1–10
Sbodio JI, Lodish HF, Chi N-W (2002) Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem J 361:451–459
Cook BD, Dynek JN, Chang W, Shostak G, Smith S (2002) Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22:332–342
Kim CA, Bowie JU (2003) SAM domains: uniform structure, diversity of function. Trends Biochem Sci 28:625–628
Qiao F, Bowie JU (2005) The many faces of SAM. Sci STKE 286:re7
De Rycker M, Price CM (2004) Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol Cell Biol 24:9802–9812
Mariotti L et al (2016) Tankyrase requires SAM domain-dependent polymerization to support Wnt-Beta-catenin signaling. Mol Cell 63:498–513
Riccio AA, McCauley M, Langelier MF, Pascal JM (2016) Tankyrase sterile alpha motif domain polymerization is required for its role in Wnt signaling. Structure 24:1573–1581
Schwarz-Romond T et al (2007) The DIX domain of dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol 14:484–492
Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z (2004) The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13:1435–1448
Li J, Mahajan A, Tsai M-D (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45:15168–15178
Seimiya H, Smith S (2002) The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J Biol Chem 277:14116–14126
Morrone S, Cheng Z, Moon RT, Cong F, Xu W (2012) Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci USA 109:1500–1505
Eisemann T et al (2016) Tankyrase-1 ankyrin repeats form an adaptable binding platform for targets of ADP-ribose modification. Structure 24:1679–1692
Sbodio JI, Chi NW (2002) Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1: NuMA contains this RXXPDG motif and is a novel tankyrase partner. J Biol Chem 277:31887–31892
Li B et al (2016) Crystal structure of a tankyrase 1–telomere repeat factor 1 complex. Acta Crystallogr F Struct Biol Commun 2:320–327
DaRosa PA, Klevit RE, Xu W (2018) Structural basis for tankyrase-RNF146 interaction reveals noncanonical tankyrase-binding motifs. Protein Sci 27:1057–1067
Bisht KK et al (2012) GDP-Mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-Ribose) polymerase activity. Mol Cell Biol 32:3044–3053
Zhou Z, Chan CH, Xiao Z, Tan E (2011) Ring finger protein 146/Iduna is a Poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adhes Migr 5:463–471
DaRosa PA et al (2014) Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 17:223–226
Wang Z et al (2012) Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev 26:235–240
Kang HC et al (2011) Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci USA 108:14103–14108
Yang L et al (2017) Tankyrase1-mediated poly(ADP-ribosyl)ation of TRF1 maintains cell survival after telomeric DNA damage. Nucleic Acids Res 45:3906–3921
Shibata A et al (2010) Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the Maintenance of G2/M checkpoint arrest. Mol Cell Biol 30:3371–3383
Panier S, Boulton SJ (2014) Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15:7–18
Huen MSY, Sy SMH, Chen J (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11:138–148
Nagy Z et al (2016) Tankyrases promote homologous recombination and check point activation in response to DSBs. PLoS Genet 12:1–26
Xu Y et al (2010) The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol 191:31–43
Shao G et al (2009) MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev 23:740–754
Feng L, Huang T, Chen J (2009) MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev 23:719–728
Naderali E et al (2018) Regulation and modulation of PTEN activity. Mol Biol Rep 45:2869–2881
Liu Q, Turner KM, Yung WKA, Chen K, Zhang W (2014) Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro Oncol 16:1313–1323
Li N et al (2015) Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev 29:157–170
Calado R, Dumitriu B (2013) Telomere dynamics in mice and humans. Semin Hematol 50:165–174
Deng Y, Chan S, Chang S (2008) Telomere dysfunction and tumor suppression-the senescence connection. Nat Rev Cancer 8:450–458
De Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19:2100–2110
Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73:177–208
Chang W, Dynek JN, Smith S (2003) TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev 17:1328–1333
Seimiya H, Muramatsu Y, Ohishi T, Tsuruo T (2005) Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell 7:25–37
Donigian JR, De Lange T (2007) The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J Biol Chem 282:22662–22667
Scherthan H et al (2000) Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell 11:4189–4203
Bailey SM, Cornforth MN (2007) Telomeres and DNA double-strand breaks: ever the twain shall meet? Cell Mol Life Sci 64:2956–2964
Bailey SM et al (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci USA 96:14899–14904
Van Steensel B, Smogorzewska A, De Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92:401–413
Dregalla RC et al (2010) Regulatory roles of tankyrase 1 at telomeres and in DNA repair: suppression of T-SCE and stabilization of DNA-pkcs. Aging (Albany NY) 2:691–708
Zou LH et al (2015) TNKS1BP1 functions in DNA double-strand break repair though facilitating DNA-PKcs autophosphorylation dependent on PARP-1. Oncotarget 6:7011–7022
Jaco I (2015) Functional interaction between poly(ADP- ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol 2:1595–1607
Peters JM, Nishiyama T (2012) Sister chromatid cohesion. Cold Spring Harb Perspect Biol 4:1–18
Sajesh BV, Lichtensztejn Z, McManus KJ (2013) Sister chromatid cohesion defects are associated with chromosome instability in Hodgkin lymphoma cells. BMC Cancer 13:391
Barber TD et al (2008) Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA 105:3443–3448
Hoque MT, Ishikawa F (2002) Cohesin defects lead to premature sister chromatid separation, kinetochore dysfunction, and spindle-assembly checkpoint activation. J Biol Chem 277:42306–42314
Hsiao SJ, Smith S (2009) Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol 184:515–526
Dynek JN (2004) Resolution of sister telomere association is required for progression through mitosis. Science 304:97–100
Kim MK, Smith S, Bloom KS (2014) Persistent telomere cohesion triggers a prolonged anaphase. Mol Biol Cell 25:30–40
Callow MG et al (2011) Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS One 6:e22595
Zhang Y et al (2011) RNF146 is a poly(ADP-ribose)directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Publ Group 13:623–629
Tripathi E, Smith S (2016) Cell cycle-regulated ubiquitination of tankyrase 1 by RNF8 and ABRO1/BRCC36 controls the timing of sister telomere resolution. EMBO J 36:1–17
Maiato H, Gomes A, Sousa F, Barisic M (2017) Mechanisms of chromosome congression during mitosis. Biology (Basel) 6:E13
Ozaki Y et al (2012) Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Mol Cell 47:694–706
Elting MW, Prakash M, Udy DB, Dumont S (2017) Mapping load-bearing in the mammalian spindle reveals local kinetochore-fiber anchorage that provides mechanical isolation and redundancy. bioRxiv 27:1–30
Chang W, Dynek JN, Smith S (2005) NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J 184:177–184
Chang P, Coughlin M, Mitchison TJ (2009) Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell 20:4575–4585
Kim G et al (2015) FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res 21:4257–4261
Scott LJ (2017) Niraparib: first global approval. Drugs 77:1029–1034
Xu D et al (2001) Telomerase activity in plasma cell dyscrasias. Br J Cancer 84:621–625
Gelmini S et al (2004) Tankyrase, a positive regulator of telomere elongation, is over expressed in human breast cancer. Cancer Lett 216:81–87
Gelmini S et al (2006) Distribution of Tankyrase-1 mRNA expression in colon cancer and its prospective correlation with progression stage. Oncol Rep 16:1261–1266
Gelmini S et al (2007) Tankytase-1 mRNA expression in bladder cancer and paired urine sediment: preliminary experience. Clin Chem Lab Med 45(7):862–866
Tang B et al (2012) Expression of TNKS1 is correlated with pathologic grade and Wnt/β-catenin pathway in human astrocytomas. J Clin Neurosci 19:139–143
Hodes R (2001) Molecular targeting of cancer: telomeres as targets. Proc Natl Acad Sci USA 98:7649–7651
Ahmed A, Tollefsbol T (2003) Telomeres, telomerase, and telomerase inhibition: clinical implications for cancer. J Am Geriatr Soc 51:116–122
Zhang HAO, Yang M, Zhao J, Chen L, Yu S (2010) Inhibition of tankyrase 1 in human gastric cancer cells enhances telomere shortening by telomerase inhibitors. Oncol Rep 24:1059–1065
Polakis P (2012) Wnt signaling in cancer. Cold Spring Harb Perspect Biol 4:1–13
Novellasdemunt L, Antas P, Li VSW (2015) Targeting Wnt signaling in colorectal cancer. Am J Physiol Cell Physiol 309:C511–C521
Renna C, Salaroli R, Cocchi C, Cenacchi G (2015) XAV939-mediated ARTD activity inhibition in human MB cell lines. PLoS One 10:1–14
Scarborough HA et al (2017) AZ1366: an inhibitor of tankyrase and the canonical Wnt pathway that limits the persistence of non-small cell lung cancer cells following EGFR inhibition. Clin Cancer Res 23:1531–1541
Thomson DW et al (2017) Discovery of a highly selective Tankyrase inhibitor displaying growth inhibition effects against a diverse range of tumor derived cell lines. J Med Chem 60:5455–5471
Lupo B et al (2016) Tankyrase inhibition impairs directional migration and invasion of lung cancer cells by affecting microtubule dynamics and polarity signals. BMC Biol 14:5
Wu X, Luo F, Li J, Zhong X, Liu K (2016) Tankyrase 1 inhibitor XAV939 increases chemosensitivity in colon cancer cell lines via inhibition of the Wnt signaling pathway. Int J Oncol 48:1333–1340
Wang H et al (2016) Tankyrase inhibitor sensitizes lung cancer cells to endothelial growth factor receptor (EGFR) inhibition via stabilizing angiomotins and inhibiting yap signaling. J Biol Chem 291:15256–15266
Arqués O et al (2016) Tankyrase inhibition blocks Wnt/b-catenin pathway and reverts resistance to PI3 K and AKT inhibitors in the treatment of colorectal cancer. Clin Cancer Res 22:644–656
Bao R et al (2012) Inhibition of tankyrases induces axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS One 7:e48670
Casás-Selves M et al (2012) Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res 72:4154–4164
Tian X et al (2013) XAV939, a tankyrase 1 inhibitior, promotes cell apoptosis in neuroblastoma cell lines by inhibiting Wnt/β-catenin signaling pathway. J Exp Clin Cancer Res 32:1–9
Stratford EW et al (2014) The tankyrase-specific inhibitor JW74 affects cell cycle progression and induces apoptosis and differentiation in osteosarcoma cell lines. Cancer Med 3:36–46
Lau T et al (2013) A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res 73:3132–3144
Mccabe N et al (2009) Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene 28:1465–1470
Kulak O et al (2015) Disruption of Wnt/beta-catenin signaling and telomeric shortening are inextricable consequences of tankyrase inhibition in human cells. Mol Cell Biol 35:2425–2435
Guo HL et al (2012) The Axin/TNKS complex interacts with KIF3A and is required for insulin-stimulated GLUT4 translocation. Cell Res 22:1246–1257
Levaot N et al (2011) 3BP2-deficient mice are osteoporotic with impaired osteoblast and osteoclast functions. J Clin Inves 131:3244–3257
Thorsell AG et al (2017) Structural basis for potency and promiscuity in poly(ADP-ribose) polymerase (PARP) and tankyrase inhibitors. J Med Chem 60:1262–1271
Coster G, Goldberg M (2010) The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus 1:166–178
Vikrant, Sawant UU,Varma AK (2014) Role of MERIT40 in stabilization of BRCA1 complex: a protein–protein interaction study. Biochem Biophys Res Commun 446:1139–1144
Acknowledgements
Pascal laboratory research on PARP enzymes is supported by grants from the Canadian Institute of Health Research (BMA342854 and PJT374609).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Eisemann, T., Pascal, J.M. Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell. Mol. Life Sci. 77, 19–33 (2020). https://doi.org/10.1007/s00018-019-03366-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03366-0