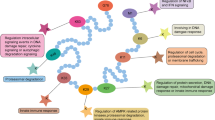
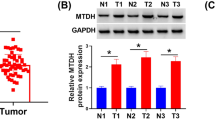
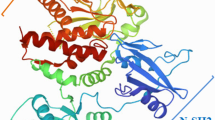
Abstract
Ubiquitin modification plays significant roles in protein fate determination, signaling transduction, and cellular processes. Over the past 2 decades, the number of studies on ubiquitination has demonstrated explosive growth. E3 ubiquitin ligases are the key enzymes that determine the substrate specificity and are involved in cancer. Several recent studies shed light on the functions and mechanisms of HECTD3 E3 ubiquitin ligase. This review describes the progress in the recent studies of HECTD3 in cancer and other diseases. We propose that HECTD3 is a potential biomarker and a therapeutic target, and discuss the future directions for HECTD3 investigations.





Similar content being viewed by others
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- ANGPT1:
-
Angiopoietin 1
- BRCA:
-
Breast invasive carcinoma
- BRCA1:
-
Breast cancer type 1 susceptibility protein
- c-Abl:
-
Abelson murine leukemia viral homolog 1
- CHOL:
-
Cholangiocarcinoma
- CRAF:
-
RAF proto-oncogene serine/threonine-protein kinase
- CRL7:
-
Cullin-RING E3 ubiquitin ligase 7
- CUL1:
-
Cullin1
- CUL7:
-
Cullin 7
- DECL:
-
DNA-encoded compound libraries
- DISC:
-
Death-inducing signaling complex
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- DUB:
-
Deubiquitinating enzyme
- E1:
-
Ubiquitin-activating enzyme
- E2:
-
Ubiquitin-conjugating enzyme
- E3:
-
Ubiquitin ligase
- E6-AP:
-
E6-associated protein
- EAE:
-
Experimental autoimmune encephalomyelitis
- ECT2:
-
Epithelial cell transforming 2
- EGFR:
-
Epidermal growth factor receptor
- ER:
-
Endoplasmic reticulum
- ERBB4:
-
Erb-b2 receptor tyrosine kinase 4
- ERK:
-
Mitogen-activated protein kinase 1
- ESCC:
-
Esophageal squamous cell carcinoma
- FBDD:
-
Fragment-based drug discovery
- FBW7:
-
F-box and WD repeat domain containing 7
- FP-HTS:
-
Fluorescence polarization assay for high-throughput screening
- HECT:
-
Homologous to E6AP C terminus
- HECTD3:
-
Homologous to the E6-associated protein carboxyl terminus domain containing 3
- HER2:
-
Erb-b2 receptor tyrosine kinase 2
- HIF1α:
-
Hypoxia inducible factor 1 subunit alpha
- HSP90:
-
Heat shock protein 90
- HTS:
-
High-throughput screening technologies
- HUWE1:
-
HECT, UBA, and WWE domain containing E3 ubiquitin protein ligase 1
- IFN:
-
Interferon
- IRE1α:
-
Inositol requiring enzyme 1 alpha
- IRF3:
-
Interferon regulatory factor 3
- ITCH:
-
Itchy E3 ubiquitin protein ligase
- KLF5:
-
Kruppel like factor 5
- LATS1:
-
Large tumor suppressor kinase 1
- LIHC:
-
Liver hepatocellular carcinoma
- LGG:
-
Brain lower grade glioma
- LUAD:
-
Lung adenocarcinoma
- MALT1:
-
MALT1 paracaspase
- MCL1:
-
Myeloid cell leukemia 1
- MDM2:
-
Murine double minute 2
- miR-153:
-
MicroRNA-153
- NEDD4-1:
-
NEDD4 E3 ubiquitin protein ligase
- OV:
-
Ovarian serous cystadenocarcinoma
- PMA:
-
Phorbol-12-myristate-13-acetate
- PML:
-
Promyelocytic leukemia protein
- PTEN:
-
Phosphatase and tensin homolog
- RBR:
-
RING-IBR-RINGs
- RING:
-
Really interesting new genes
- RLD:
-
RCC1 like domain
- RNF20:
-
Ring finger protein 20
- RORγt:
-
Retineic-acid-receptor-related orphan nuclear receptor γ
- SCF:
-
SKP1-CUL1-F-box protein
- SKP2:
-
S-phase kinase-associated protein 2
- SMAD2:
-
SMAD family member 2
- SMURF2:
-
SMAD specific E3 ubiquitin protein ligase 2
- Stat3:
-
Signal transducer and activator of transcription 3
- Tara:
-
Trio-associated repeat on actin
- TBK1:
-
TANK binding kinase 1
- TGFβ:
-
Transforming growth factor β
- TGFβR1:
-
Transforming growth factor β receptor 1
- Th17:
-
T helper 17
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TNBC:
-
Triple negative breast cancer
- TRAF3:
-
TNF receptor-associated factor 3
- TRAF6:
-
TNF receptor-associated factor 6
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- Ub:
-
Ubiquitin
- UCEC:
-
Uterine Corpus Endometrial Carcinoma
- UCS:
-
Uterine Carcinosarcoma
- UbV:
-
Ub variant
- UCS:
-
Uterine carcinosarcoma
- VCB-CR:
-
pVHL-elongin C-elongin B-cullin 2-RBX1
- VHL:
-
Von Hippel–Lindau disease tumor suppressor
- WWP1:
-
WW domain containing E3 ubiquitin protein ligase 1
- WWP2:
-
WW domain containing E3 ubiquitin protein ligase 2
- XBP1:
-
X-box binding protein 1
References
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Rape M (2018) Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol 19(1):59–70. https://doi.org/10.1038/nrm.2017.83
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229. https://doi.org/10.1146/annurev-biochem-060310-170328
Tokunaga F, Sakata S-i, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11:123. https://doi.org/10.1038/ncb1821. https://www.nature.com/articles/ncb1821#supplementary-information
Trempe JF (2011) Reading the ubiquitin postal code. Curr Opin Struct Biol 21(6):792–801. https://doi.org/10.1016/j.sbi.2011.09.009
Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10(6):398–409. https://doi.org/10.1038/nrm2690
Haglund K, Dikic I (2005) Ubiquitylation and cell signaling. EMBO J 24(19):3353–3359. https://doi.org/10.1038/sj.emboj.7600808
Hoeller D, Hecker CM, Dikic I (2006) Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer 6(10):776–788. https://doi.org/10.1038/nrc1994
Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science (New York, NY) 315(5809):201–205. https://doi.org/10.1126/science.1127085
Huen MS, Sy SM, Chen J (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11(2):138–148. https://doi.org/10.1038/nrm2831
Vucic D, Dixit VM, Wertz IE (2011) Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol 12(7):439–452. https://doi.org/10.1038/nrm3143
Gilberto S, Peter M (2017) Dynamic ubiquitin signaling in cell cycle regulation. J Cell Biol 216(8):2259–2271. https://doi.org/10.1083/jcb.201703170
Senft D, Qi J, Ronai ZA (2018) Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer 18(2):69–88. https://doi.org/10.1038/nrc.2017.105
Popovic D, Vucic D, Dikic I (2014) Ubiquitination in disease pathogenesis and treatment. Nat Med 20(11):1242–1253. https://doi.org/10.1038/nm.3739
He M, Zhou Z, Wu G, Chen Q, Wan Y (2017) Emerging role of DUBs in tumor metastasis and apoptosis: therapeutic implication. Pharmacol Ther 177:96–107
Harrigan JA, Jacq X, Martin NM, Jackson SP (2018) Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov 17(1):57–78. https://doi.org/10.1038/nrd.2017.152
Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK (2012) The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149(5):1098–1111. https://doi.org/10.1016/j.cell.2012.02.065
Dubrez L, Rajalingam K (2015) IAPs and cell migration. Semin Cell Dev Biol 39:124–131. https://doi.org/10.1016/j.semcdb.2015.02.015
Kim H, Frederick DT, Levesque MP, Cooper ZA, Feng Y, Krepler C, Brill L, Samuels Y, Hayward NK, Perlina A, Piris A, Zhang T, Halaban R, Herlyn MM, Brown KM, Wargo JA, Dummer R, Flaherty KT, Ronai ZA (2015) Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep 11(9):1458–1473. https://doi.org/10.1016/j.celrep.2015.04.049
Randle SJ, Laman H (2016) F-box protein interactions with the hallmark pathways in cancer. Semin Cancer Biol 36:3–17. https://doi.org/10.1016/j.semcancer.2015.09.013
Yang L, Chen J, Huang X, Zhang E, He J, Cai Z (2018) Novel insights Into E3 ubiquitin ligase in cancer chemoresistance. Am J Med Sci 355(4):368–376. https://doi.org/10.1016/j.amjms.2017.12.012
Buetow L, Huang DT (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 17(10):626–642. https://doi.org/10.1038/nrm.2016.91
Wade M, Li Y-C, Wahl GM (2013) MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13(2):83–96. https://doi.org/10.1038/nrc3430
Frescas D, Pagano M (2008) Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 8(6):438–449. https://doi.org/10.1038/nrc2396
Xu W, Taranets L, Popov N (2016) Regulating Fbw7 on the road to cancer. Semin Cancer Biol 36:62–70. https://doi.org/10.1016/j.semcancer.2015.09.005
Gossage L, Eisen T, Maher ER (2015) VHL, the story of a tumour suppressor gene. Nat Rev Cancer 15(1):55–64. https://doi.org/10.1038/nrc3844
Li ML, Greenberg RA (2012) Links between genome integrity and BRCA1 tumor suppression. Trends Biochem Sci 37(10):418–424. https://doi.org/10.1016/j.tibs.2012.06.007
Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barrière C, Stuckey JA, Meagher JL, Bai L, Liu L, Hoffman-Luca CG, Lu J, Shangary S, Yu S, Bernard D, Aguilar A, Dos-Santos O, Besret L, Guerif S, Pannier P, Gorge-Bernat D, Debussche L (2014) SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Can Res 74(20):5855–5865. https://doi.org/10.1158/0008-5472.Can-14-0799
Aguilar A, Lu J, Liu L, Du D, Bernard D, McEachern D, Przybranowski S, Li X, Luo R, Wen B, Sun D, Wang H, Wen J, Wang G, Zhai Y, Guo M, Yang D, Wang S (2017) Discovery of 4-((3′R,4′S,5′R)-6″-Chloro-4′-(3-chloro-2-fluorophenyl)-1′-ethyl-2″-oxodispiro[cyclohexane-1,2′-pyrrolidine-3′,3″-indoline]-5′-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): a potent and orally active murine double minute 2 (MDM2) inhibitor in clinical development. J Med Chem 60 (7):2819–2839. https://doi.org/10.1021/acs.jmedchem.6b01665
So WV, Ou Yang T-H, Yang X, Zhi J (2019) Lack of UGT polymorphism association with idasanutlin pharmacokinetics in solid tumor patients. Cancer Chemother Pharmacol 83(1):209–213. https://doi.org/10.1007/s00280-018-3741-2
Sluimer J, Distel B (2018) Regulating the human HECT E3 ligases. Cell Mol Life Sci 75(17):3121–3141. https://doi.org/10.1007/s00018-018-2848-2
Zheng N, Shabek N (2017) Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem 86:129–157. https://doi.org/10.1146/annurev-biochem-060815-014922
Wenzel DM, Lissounov A, Brzovic PS, Klevit RE (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474(7349):105–108. https://doi.org/10.1038/nature09966
Lorenz S (2018) Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem 399(2):127–145. https://doi.org/10.1515/hsz-2017-0184
Scheffner M, Nuber U, Huibregtse JM (1995) Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature 373(6509):81–83
Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (1999) Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science (New York, NY) 286(5443):1321–1326
Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92(11):5249
Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75(3):495–505
Louria-Hayon I, Alsheich-Bartok O, Levav-Cohen Y, Silberman I, Berger M, Grossman T, Matentzoglu K, Jiang YH, Muller S, Scheffner M, Haupt S, Haupt Y (2009) E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ 16(8):1156–1166. https://doi.org/10.1038/cdd.2009.31
Wolyniec K, Shortt J, de Stanchina E, Levav-Cohen Y, Alsheich-Bartok O, Louria-Hayon I, Corneille V, Kumar B, Woods SJ, Opat S, Johnstone RW, Scott CL, Segal D, Pandolfi PP, Fox S, Strasser A, Jiang YH, Lowe SW, Haupt S, Haupt Y (2012) E6AP ubiquitin ligase regulates PML-induced senescence in Myc-driven lymphomagenesis. Blood 120(4):822–832. https://doi.org/10.1182/blood-2011-10-387647
Paul PJ, Raghu D, Chan AL, Gulati T, Lambeth L, Takano E, Herold MJ, Hagekyriakou J, Vessella RL, Fedele C, Shackleton M, Williams ED, Fox S, Williams S, Haupt S, Gamell C, Haupt Y (2016) Restoration of tumor suppression in prostate cancer by targeting the E3 ligase E6AP. Oncogene 35(48):6235–6245. https://doi.org/10.1038/onc.2016.159
Mansour M, Haupt S, Chan AL, Godde N, Rizzitelli A, Loi S, Caramia F, Deb S, Takano EA, Bishton M, Johnstone C, Monahan B, Levav-Cohen Y, Jiang YH, Yap AS, Fox S, Bernard O, Anderson R, Haupt Y (2016) The E3-ligase E6AP represses breast cancer metastasis via regulation of ECT2-Rho signaling. Can Res 76(14):4236–4248. https://doi.org/10.1158/0008-5472.Can-15-1553
Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W, Zhou Z, Petros J, Frierson HF, Vessella RL, Atfi A, Dong JT (2007) Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene 26(16):2386–2394
Chen C, Zhou Z, Ross JS, Zhou W, Dong JT (2007) The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer 121(1):80–87
Li Y, Zhou Z, Chen C (2008) WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ 15(12):1941–1951. https://doi.org/10.1038/cdd.2008.134
Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT (2005) Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem 280(50):41553–41561
Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K (2004) Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23(41):6914–6923. https://doi.org/10.1038/sj.onc.1207885
Li Y, Zhou Z, Alimandi M, Chen C (2009) WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer. Oncogene 28(33):2948–2958. https://doi.org/10.1038/onc.2009.162
Salah Z, Melino G, Aqeilan RI (2011) Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Can Res 71(5):2010–2020. https://doi.org/10.1158/0008-5472.Can-10-3516
Chang L, Shen L, Zhou H, Gao J, Pan H, Zheng L, Armstrong B, Peng Y, Peng G, Zhou BP, Rosen ST, Shen B (2019) ITCH nuclear translocation and H1.2 polyubiquitination negatively regulate the DNA damage response. Nucleic Acids Res 47(2):824–842. https://doi.org/10.1093/nar/gky1199
Lin X, Liang M, Feng XH (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem 275(47):36818–36822
Tang LY, Yamashita M, Coussens NP, Tang Y, Wang XC, Li CL, Deng CX, Cheng SY, Zhang YE (2011) Ablation of Smurf2 reveals an inhibition in TGF-beta signalling through multiple mono-ubiquitination of Smad3. EMBO J 30(23):4777–4789. https://doi.org/10.1038/emboj.2011.393
Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zhang YE (2012) A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat Med 18(2):227–234. https://doi.org/10.1038/nm.2596
Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, Jiang X (2007) NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128(1):129–139
Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, Chen J (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol 13(6):728–733. https://doi.org/10.1038/ncb2240
Yu J, Lan J, Zhu Y, Li X, Lai X, Xue Y, Jin C, Huang H (2008) The E3 ubiquitin ligase HECTD3 regulates ubiquitination and degradation of Tara. Biochem Biophys Res Commun 367(4):805–812. https://doi.org/10.1016/j.bbrc.2008.01.022
Zhang L, Kang L, Bond W, Zhang N (2009) Interaction between syntaxin 8 and HECTd3, a HECT domain ligase. Cell Mol Neurobiol 29(1):115–121. https://doi.org/10.1007/s10571-008-9303-0
Li Y, Chen X, Wang Z, Zhao D, Chen H, Chen W, Zhou Z, Zhang J, Zhang J, Li H, Chen C (2013) The HECTD3 E3 ubiquitin ligase suppresses cisplatin-induced apoptosis via stabilizing MALT1. Neoplasia 15(1):39-IN15. https://doi.org/10.1593/neo.121362
Cho JJ, Xu Z, Parthasarathy U, Drashansky TT, Helm EY, Zuniga AN, Lorentsen KJ, Mansouri S, Cho JY, Edelmann MJ, Duong DM, Gehring T, Seeholzer T, Krappmann D, Uddin MN, Califano D, Wang RL, Jin L, Li H, Lv D, Zhou D, Zhou L, Avram D (2019) Hectd3 promotes pathogenic Th17 lineage through Stat3 activation and Malt1 signaling in neuroinflammation. Nat Commun 10(1):701. https://doi.org/10.1038/s41467-019-08605-3
Li Y, Kong Y, Zhou Z, Chen H, Wang Z, Hsieh YC, Zhao D, Zhi X, Huang J, Zhang J, Li H, Chen C (2013) The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell Death Dis 4:e935. https://doi.org/10.1038/cddis.2013.464
Li Y, Wu X, Li L, Liu Y, Xu C, Su D, Liu Z (2017) The E3 ligase HECTD3 promotes esophageal squamous cell carcinoma (ESCC) growth and cell survival through targeting and inhibiting caspase-9 activation. Cancer Lett 404:44–52. https://doi.org/10.1016/j.canlet.2017.07.004
Li Z, Zhou L, Prodromou C, Savic V, Pearl LH (2017) HECTD3 mediates an HSP90-dependent degradation pathway for protein kinase clients. Cell Rep 19(12):2515–2528. https://doi.org/10.1016/j.celrep.2017.05.078
Li F, Li Y, Liang H, Xu T, Kong Y, Huang M, Xiao J, Chen X, Xia H, Wu Y, Zhou Z, Guo X, Hu C, Yang C, Cheng X, Chen C, Qi X (2018) HECTD3 mediates TRAF3 polyubiquitination and type I interferon induction during bacterial infection. J Clin Invest 128(9):4148–4162. https://doi.org/10.1172/JCI120406
Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D (2007) Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J 26(22):4634–4645
Kong Y, Wang Z, Huang M, Zhou Z, Li Y, Miao H, Wan X, Huang J, Mao X, Chen C (2019) CUL7 promotes cancer cell survival through promoting Caspase-8 ubiquitination. Int J Cancer. https://doi.org/10.1002/ijc.32239
Shu T, Li Y, Wu X, Li B, Liu Z (2017) Down-regulation of HECTD3 by HER2 inhibition makes serous ovarian cancer cells sensitive to platinum treatment. Cancer Lett 411:65–73. https://doi.org/10.1016/j.canlet.2017.09.048
Yu H, Jove R (2004) The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4(2):97–105. https://doi.org/10.1038/nrc1275
Wu X, Li L, Li Y, Liu Z (2016) MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. Am J Cancer Res 6(7):1563–1571
Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, Chen H, Dong C, Yang R, Liu S, Chen C (2016) Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics 6(4):533–544. https://doi.org/10.7150/thno.14315
Liang H, Xiao J, Zhou Z, Wu J, Ge F, Li Z, Zhang H, Sun J, Li F, Liu R, Chen C (2018) Hypoxia induces miR-153 through the IRE1alpha-XBP1 pathway to fine tune the HIF1alpha/VEGFA axis in breast cancer angiogenesis. Oncogene 37(15):1961–1975. https://doi.org/10.1038/s41388-017-0089-8
Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, Chen C (2018) miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis 21(4):849–860. https://doi.org/10.1007/s10456-018-9630-9
Ekambaram P, Lee JL, Hubel NE, Hu D, Yerneni S, Campbell PG, Pollock N, Klei LR, Concel VJ, Delekta PC, Chinnaiyan AM, Tomlins SA, Rhodes DR, Priedigkeit N, Lee AV, Oesterreich S, McAllister-Lucas LM, Lucas PC (2018) The CARMA3-Bcl10-MALT1 signalosome drives NFκB activation and promotes aggressiveness in angiotensin II receptor-positive breast cancer. Can Res 78(5):1225–1240. https://doi.org/10.1158/0008-5472.Can-17-1089
Cheng L, Deng N, Yang N, Zhao X, Lin X (2019) Malt1 protease is critical in maintaining function of regulatory T cells and may be a therapeutic target for antitumor immunity. J Immunol (Baltimore, Md: 1950) 202(10):3008–3019. https://doi.org/10.4049/jimmunol.1801614
Kawadler H, Gantz MA, Riley JL, Yang X (2008) The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol Cell 31(3):415–421. https://doi.org/10.1016/j.molcel.2008.06.008
Brüstle A, Brenner D, Knobbe CB, Lang PA, Virtanen C, Hershenfield BM, Reardon C, Lacher SM, Ruland J, Ohashi PS, Mak TW (2012) The NF-κB regulator MALT1 determines the encephalitogenic potential of Th17 cells. J Clin Invest 122(12):4698–4709. https://doi.org/10.1172/jci63528
Paramore A, Frantz S (2003) Bortezomib. Nat Rev Drug Discov 2(8):611–612
Adams J (2004) The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5(5):417–421. https://doi.org/10.1016/S1535-6108(04)00120-5
Qi J, Ronai ZA (2015) Dysregulation of ubiquitin ligases in cancer. Drug Resist Updat 23:1–11. https://doi.org/10.1016/j.drup.2015.09.001
Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LGGC, Masucci M, Pramanik A, Selivanova G (2004) Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat Med 10(12):1321–1328. https://doi.org/10.1038/nm1146
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, NY) 303(5659):844–848
Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, Weissman AM, Vousden KH (2005) Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 7(6):547–559
Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, Geho D, Middleton SA, Vassilev LT, Nichols GL, Bui BN (2012) Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 13(11):1133–1140. https://doi.org/10.1016/s1470-2045(12)70474-6
Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yang D, Wang S (2008) Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA 105(10):3933–3938. https://doi.org/10.1073/pnas.0708917105
Azmi AS, Aboukameel A, Banerjee S, Wang Z, Mohammad M, Wu J, Wang S, Yang D, Philip PA, Sarkar FH, Mohammad RM (2010) MDM2 inhibitor MI-319 in combination with cisplatin is an effective treatment for pancreatic cancer independent of p53 function. Eur J Cancer (Oxford, England: 1990) 46(6):1122–1131. https://doi.org/10.1016/j.ejca.2010.01.015
Zhao Y, Liu L, Sun W, Lu J, McEachern D, Li X, Yu S, Bernard D, Ochsenbein P, Ferey V, Carry JC, Deschamps JR, Sun D, Wang S (2013) Diastereomeric spirooxindoles as highly potent and efficacious MDM2 inhibitors. J Am Chem Soc 135(19):7223–7234. https://doi.org/10.1021/ja3125417
Zhao Y, Yu S, Sun W, Liu L, Lu J, McEachern D, Shargary S, Bernard D, Li X, Zhao T, Zou P, Sun D, Wang S (2013) A potent small-molecule inhibitor of the MDM2-p53 interaction (MI-888) achieved complete and durable tumor regression in mice. J Med Chem 56(13):5553–5561. https://doi.org/10.1021/jm4005708
Lu J, McEachern D, Li S, Ellis MJ, Wang S (2016) Reactivation of p53 by MDM2 Inhibitor MI-77301 for the treatment of endocrine-resistant breast cancer. Mol Cancer Ther 15(12):2887–2893
Nör F, Warner KA, Zhang Z, Acasigua GA, Pearson AT, Kerk SA, Helman JI, Sant’Ana Filho M, Wang S, Nör JE (2017) Therapeutic Inhibition of the MDM2-p53 Interaction Prevents Recurrence of Adenoid Cystic Carcinomas. Clin Cancer Res 23(4):1036–1048. https://doi.org/10.1158/1078-0432.Ccr-16-1235
Andrews A, Warner K, Rodriguez-Ramirez C, Pearson AT, Nör F, Zhang Z, Kerk S, Kulkarni A, Helman JI, Brenner JC, Wicha MS, Wang S, Nör JE (2019) Ablation of cancer stem cells by therapeutic inhibition of the MDM2-p53 interaction in mucoepidermoid carcinoma. Clin Cancer Res 25(5):1588–1600. https://doi.org/10.1158/1078-0432.Ccr-17-2730
Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, Chu XJ, Bartkovitz D, Podlaski F, Janson C, Tovar C, Filipovic ZM, Higgins B, Glenn K, Packman K, Vassilev LT, Graves B (2013) Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem 56(14):5979–5983. https://doi.org/10.1021/jm400487c
Zhang W, Wu KP, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, Mercredi PY, Murchie R, Hu J, Persaud A, Mukherjee M, Li N, Doye A, Walker JR, Sheng Y, Hao Z, Li Y, Brown KR, Lemichez E, Chen J, Tong Y, Harper JW, Moffat J, Rotin D, Schulman BA, Sidhu SS (2016) System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol Cell 62(1):121–136. https://doi.org/10.1016/j.molcel.2016.02.005
Gorelik M, Orlicky S, Sartori MA, Tang X, Marcon E, Kurinov I, Greenblatt JF, Tyers M, Moffat J, Sicheri F, Sidhu SS (2016) Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1–F-box interface. Proc Natl Acad Sci 113(13):3527. https://doi.org/10.1073/pnas.1519389113
Gabrielsen M, Buetow L, Nakasone MA, Ahmed SF, Sibbet GJ, Smith BO, Zhang W, Sidhu SS, Huang DT (2017) A general strategy for discovery of inhibitors and activators of RING and U-box E3 ligases with ubiquitin variants. Mol Cell 68(2):456.e410–470.e410
Mund T, Lewis MJ, Maslen S, Pelham HR (2014) Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci USA 111(47):16736–16741. https://doi.org/10.1073/pnas.1412152111
Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G, Bernassola F, Cavasotto CN, Knight RA, Ciechanover A, Melino G (2014) High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 5(5):e1203. https://doi.org/10.1038/cddis.2014.113
Kathman SG, Span I, Smith AT, Xu Z, Zhan J, Rosenzweig AC, Statsyuk AV (2015) A small molecule that switches a ubiquitin ligase from a processive to a distributive enzymatic mechanism. J Am Chem Soc 137(39):12442–12445. https://doi.org/10.1021/jacs.5b06839
Sander B, Xu W, Eilers M, Popov N, Lorenz S (2017) A conformational switch regulates the ubiquitin ligase HUWE1. eLife 6. https://doi.org/10.7554/elife.21036
Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, Devreotes P, Wolberger C, Amzel LM, Gabelli SB, Cole PA (2017) A tunable brake for HECT ubiquitin ligases. Mol Cell 66(3):345.e346–357.e346
Chan AL, Grossman T, Zuckerman V, Campigli Di Giammartino D, Moshel O, Scheffner M, Monahan B, Pilling P, Jiang YH, Haupt S, Schueler-Furman O, Haupt Y (2013) c-Abl phosphorylates E6AP and regulates its E3 ubiquitin ligase activity. Biochemistry 52(18):3119–3129. https://doi.org/10.1021/bi301710c
Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Church WB, Dastmalchi S (2013) Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov Today 18(23–24):1144–1157. https://doi.org/10.1016/j.drudis.2013.09.001
Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X (2013) High-throughput screening alphascreen assay for identification of small-molecule inhibitors of ubiquitin E3 Ligase SCFSkp2-Cks1. J Biomol Screen 18(8):910–920. https://doi.org/10.1177/1087057113485789
Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H (2016) Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov 15(9):605–619. https://doi.org/10.1038/nrd.2016.109
Gu L, Zhang H, Liu T, Zhou S, Du Y, Xiong J, Yi S, Qu CK, Fu H, Zhou M (2016) Discovery of dual inhibitors of MDM2 and XIAP for cancer treatment. Cancer Cell 30(4):623–636
Krist DT, Park S, Boneh GH, Rice SE, Statsyuk AV (2016) UbFluor: a mechanism-based probe for HECT E3 ligases. Chem Sci 7(8):5587–5595. https://doi.org/10.1039/C6SC01167E
Veggiani G, Gerpe MCR, Sidhu SS, Zhang W (2019) Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol Ther
Franzini RM, Neri D, Scheuermann J (2014) DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries. Acc Chem Res 47(4):1247–1255. https://doi.org/10.1021/ar400284t
Rognan D (2017) The impact of in silico screening in the discovery of novel and safer drug candidates. Pharmacol Ther 175:47–66
Li Y, Xie P, Lu L, Wang J, Diao L, Liu Z, Guo F, He Y, Liu Y, Huang Q, Liang H, Li D, He F (2017) An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat Commun 8(1):347. https://doi.org/10.1038/s41467-017-00299-9
Herman AG, Hayano M, Poyurovsky MV, Shimada K, Skouta R, Prives C, Stockwell BR (2011) Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov 1(4):312–325. https://doi.org/10.1158/2159-8290.Cd-11-0104
Tian M, Zeng T, Liu M, Han S, Lin H, Lin Q, Li L, Jiang T, Li G, Lin H, Zhang T, Kang Q, Deng X, Wang H-R (2019) A cell-based high-throughput screening method based on a ubiquitin-reference technique for identifying modulators of E3 ligases. J Biol Chem 294(8):2880–2891. https://doi.org/10.1074/jbc.ra118.003822
Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res 12(1):260–271. https://doi.org/10.1021/pr300630k
Acknowledgements
This study was supported in part by grants from the National Key R&D Program of China (2018YFC2000400) and the National Nature Science Foundation of China (81830087, U1602221, and 31771516 to Chen, C and 81773149 to Kong Y) and the Shenzhen Municipal Government of China (KQTD20170810160226082).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, Q., Li, F., Cheng, Z. et al. The role of E3 ubiquitin ligase HECTD3 in cancer and beyond. Cell. Mol. Life Sci. 77, 1483–1495 (2020). https://doi.org/10.1007/s00018-019-03339-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03339-3