Summary
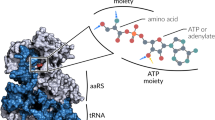
The fidelity of protein biosynthesis rests not only on the proper interaction of the messenger RNA codon with the anticodon of the tRNA, but also on the correct attachment of amino acids to their corresponding (cognate) transfer RNA (tRNA) species. This process is catalyzed by the aminoacyl-tRNA synthetases which discriminate with remarkable selectivity amongst many structurally similar tRNAs. The basis for this highly specific recognition of tRNA by these enzymes (also referred to as ‘tRNA identity’) is currently being elucidated by genetic, biochemical and biophysical techniques. At least two factors are important in determining the accuracy of aminoacylation: a) ‘identity elements’ in tRNA denote nucleotides in certain positions crucial for protein interactions determining specificity, and b) the occurrence in vivo of competition between synthetases for a particular tRNA which may have ambiguous identity.
Similar content being viewed by others
References
Akins, A., and Lambowitz, A., A protein required for splicing group I introns inNeurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell50 (1987) 331–345.
Baldwin, A. N., and Berg, P., Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. biol. Chem.241 (1966) 839–845.
Barat, C., Lullien, V., Schatz, O., Keith, G., Nugeyre, M. T., Gruninger-Leitch, F., Barre-Sinoussi, F., LeGrice, S. F., and Darlix, J. L., HIV-1 reverse transcriptase specifically interacts with the anti-codon domain of its cognate primer tRNA. EMBO J.8 (1989) 3279–3285.
Bedouelle, H., and Winter, G., A model of synthetase/transfer RNA interaction as deduced by protein engineering. Nature320 (1986) 371–373.
Brick, P., Bhat, T. N., and Blow, D. M., Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution. Interaction of the enzyme with tyrosyl adenylate intermediate. J. molec. Biol.208 (1989) 83–98.
Crothers, D. M., Seno, T., and Söll, D., Is there a discriminator site in transfer RNA? Proc. natl Acad. Sci. USA69 (1972) 3063–3067.
deDuve, C., Transfer RNAs: the second genetic code. Nature333 (1988) 117–118.
Eggertsson, G., and Söll, D., Transfer RNA-mediated suppression of termination codons inEscherichia coli. Microbiol. Rev.52 (1988) 354–374.
Eriani, G., Delarue, M., Poch, O., Gangloff, J., and Moras, D., Primary sequence of ProRS and partition of all tRNA-synthetases into two classes, as revealed by three new sequence motifs. Nature347 (1990) 203–206.
Ferguson, B. Q., and Yang, D. C., Methionyl-tRNA synthetase induced 3′-terminal and delocalized conformational transition in tRNAfMet: steady-state fluorescence of tRNA with a single fluorophore. Biochemistry25 (1986) 529–539.
Freist, W., Mechanisms of aminoacyl-tRNA synthetases: a critical consideration of recent results. Biochemistry28 (1989) 6787–6795.
Ghysen, A., and Celis, J. E., Mischarging single and double mutants ofEscherichia coli su3 tyrosine transfer RNA. J. molec. Biol.83 (1974) 333–351.
Hartman, P. E., and Roth, J. R., Mechanisms of suppression. Adv. Genet.17 (1973) 1–105.
Hecht, S. M., 2′-OH vs 3′-OH specificity in tRNA aminoacylation, in: Transfer RNA: Structure, Properties and Recognition, pp. 345–360. Eds P. R. Schimmel, D. Söll, and J. N. Abelson. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. 1979.
Hooper, J. L., Russell, R. L., and Smith, J. D., Mischarging in mutant tyrosine transfer RNAs. FEBS Lett.22 (1972) 149–155.
Hou, Y.-M., and Schimmel, P., A simple structural feature is a major determinant of the identity of a transfer RNA. Nature333 (1988) 140–145.
Hou, Y.-M., and Schimmel, P., Modeling with in vitro parameters for the elaboration of transfer RNA identity in vitro. Biochemistry28 (1989) 4942–4947.
Inokuchi, H., Celis, J. E., and Smith, J. D., Mutant tyrosine transfer ribonucleic acids ofEscherichia coli: construction by recombination of a double mutant A1G82 chargeable with glutamine. J. molec. Biol.85 (1974) 187–192.
Inokuchi, H., Hoben, P., Yamao, F., Ozeki, H., and Söll, D., Transfer RNA mischarging mediated by a mutantEscherichia coli glutaminyl-tRNA synthetase. Proc. natl Acad. Sci. USA81 (1984) 5076–5080.
Jakubowski, H., and Goldman, E., Quantities of individual aminoacyl-tRNA families and their turnover inEscherichia coli. J. Bact.158 (1984) 769–776.
Jasin, M., Regan, L., and Schimmel, P., Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature, Lond.306 (1983) 441–447.
Kim, S. H., Crystal structure of yeast tRNAPhe and general structural features of other tRNAs, in: Transfer RNA: Structure, Properties and Recognition, pp. 83–100. Eds P. R. Schimmel, D. Söll and J. N. Abelson. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. 1979.
Komine, Y., Adachi, T., Inokuchi, H., and Ozeki, H., Genomic organization and physical mapping of the transfer RNA genes inEscherichia coli K12. J. molec. Biol.212 (1990) 579–598.
Leatherbarrow, R. J., and Fersht, A. R., Protein Engineering. Protein Engineering1 (1986) 7–16.
Loftfield, R. B., and Vanderjagt, M. A., The frequency of errors in protein biosynthesis. Biochem. J.128 (1972) 1353–1356.
McClain, W. H., and Foss, K., Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science240 (1988) 793–796.
McClain, W. H., and Nicholas, H. B. Jr, Differences between transfer RNA molecules. J. molec. Biol.194 (1987) 635–642.
Monteilhet, C., and Blow, D. M., Binding of tyrosine, adenosine triphosphate and analogues to crystalline tyrosyl transfer RNA synthetase. J. molec. Biol.122 (1978) 407–417.
Muramatsu, T., Nishikawa, K., Nemoto, F., Kuchino, Y., Nishimura, S., Miyazawa, T., and Yokoyama, S., Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature336 (1988) 179–181.
Murgola, E. J., tRNA, suppression, and the code. A. Rev. Genet.19 (1985) 57–80.
Normanly, J., and Abelson, J., Transfer RNA identity. A. Rev. Biochem.58 (1989) 1029–1049.
Normanly, J., Ogden, R. C., Horvath, S. J., and Abelson, J., Changing the identity of a transfer RNA. Nature321 (1986) 213–219.
O'Connor, M., Gesteland, R. F., and Atkins, J. F., tRNA hopping: enhancement by an expanded anticodon. EMBO J.8 (1989) 4315–4323.
Ozeki, H., Inokuchi, H., Yamao, F., Kodaira, M., Sakano, H., Ikemura, T., and Shimura, Y., Genetics of nonsense suppressor tRNAs inEscherichia coli, in: Transfer RNA: Biological Aspects, pp. 341–362. Eds D. Söll, J. N. Abelson and P. R. Schimmel. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. 1980.
Perona, J. J., Swanson, R. N., Rould, M. A., Steitz, T. A., and Söll, D., Structural basis for misaminoacylation by mutantE. coli glutaminyl-tRNA synthetase enzymes. Science246 (1989) 1152–1154.
Perona, J. J., Swanson, R., Steitz, T. A., and Söll, D., Overproduction and purification ofEscherichia coli tRNA Gln2 and its use in crystallization of the glutaminyl-tRNA synthetase: tRNAGln complex. J. molec. Biol.202 (1988) 121–126.
Remme, J., Margus, T., Villems, R., and Nierhaus, K. H., The third ribosomal transfer RNA-binding site, the E site, is occupied in native polysomes. Eur. J. Biochem.183 (1989) 281–284.
Rogers, M. J., and Söll, D., Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc. natl Acad. Sci. USA85 (1988) 6627–6631.
Rouget, P., and Chapeville, F., Leucyl-tRNA synthetase. Two forms of the enzyme: relation between structural and catalytic properties. Eur. J. Biochem.23 (1971) 459–467.
Rould, M. A., Perona, J., Söll, D., and Steitz, T., Structure ofE. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science246 (1989) 1135–1142.
Rubin, J., and Blow, D. M., Amino acid activation in crystalline tyrosyl-tRNA synthetase fromBacillus stearothermophilus. J. molec. Biol.145 (1981) 489–500.
Sampson, J. R., DiRenzo, A. B., Behlen, L. S., and Uhlenbeck, O. C., Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science243 (1989) 1363–1366.
Sampson, J. R., and Uhlenbeck, O. C., Biochemical and physical characterization of an unmodified yeast phenylalamine transfer RNA transcribed in vitro. Proc. natl Acad. Sci.85 (1988) 1033–1037.
Schimmel, P., Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of tRNAs. A. Rev. Biochem.56 (1987) 125–158.
Schön, A., Kannangara, C. G., Gough, S., and Söll, D., Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature331 (1988) 187–190.
Schön, A., Krupp, G., Gough, S., Berry-Lowe, S., Kannangara, C. G., and Söll, D., The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature, Lond.322 (1986) 281–284.
Schulman, L. H., and Pelka, H., In vitro conversion of a methionine to a glutamine acceptor tRNA. Biochemistry24 (1985) 7309–7314.
Schulman, L. H., and Pelka, H., Anticodon switching changes the identity of methionine and valine transfer RNAs. Science242 (1988) 765–768.
Seno, T., Agris, P. F., and Söll, D., Involvement of the anticodon region ofEscherichia coli tRNAGln and tRNAGlu in the specific interaction with cognate aminoacyl tRNA synthetase: Alteration of the 2-thiouridine derivitives in the anticodon of the tRNAs by BrCN or sulfur deprivation. Biochim. biophys. Acta349 (1974) 328–338.
Shimura, Y., Aono, H., Ozeki, H., Sarabhai, A., Lamfrom, H., and Abelson, J., Mutant tyrosine tRNA of altered amino acid specificity. FEBS Lett.22 (1972) 144–148.
Shoffner, J. M., Lott, M. T., Lezza, A. M. S., Seibel, P., Ballinger, S. W., and Wallace, D. C., Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell61 (1990) 931–937.
Smith, J. D., and Celis, J. E., Mutant tyrosine transfer RNA that can be charged with glutamine. Nature New Biol.243 (1973) 66–71.
Söll, D., and Schimmel, P. R., Aminoacyl-tRNA synthetases, in: The Enzymes, vol. X, pp. 489–538. Ed. P. Boyer. Academic Press, San Francisco 1974.
Sprinzl, M., Hartmann, T., Meissner, F., Moll, J., and Vorderwülbecke, T., Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res.15 (1987) r53–r188.
Swanson, R., Hoben, P., Sumner-Smith, M., Uemura, H., Watson, L., and Söll, D., Accuracy of in vivo aminoacylation requires the proper balance of tRNA and aminoacyl-tRNA synthetase. Science242 (1988) 1548–1551.
Thorbjarnardottir, S., Dingermann, T., Rafnar, T., Andresson, O.S., Söll, D., and Eggertsson, G., Leucine tRNA family ofEscherichia coli: nucleotide sequence of the supP(Am) suppressor gene. J. Bact.161 (1985) 219–222.
Uemura, H., Conley, J., Yamao, F., Rogers, J., and Söll, D.,Escherichia coli glutaminyl-tRNA synthetase: a single aminoa cid replacement relaxes tRNA specificity. Protein Sequences Data Analysis1 (1988) 479–485.
Uemura, H., Rogers, M.J., Swanson, R., Watson, L., and Söll, D., Site-directed mutagenesis to fine-tune enzyme specifity. Protein Engineering2 (1988) 293–296.
Waldrop, M. M., The structure of the ‘second genetic code’. Science246 (1989) 1122.
Williamson, R. M., and Oxender, D. L., Sequence and structural similarities between the leucine-specific binding protein and leucyl-tRNA synthetase ofEscherichia coli. Proc. natl Acad. Sci. USA87 (1990) 4561–4565.
Yaniv, M., Folk, W. R., Berg, P., and Soll, L., A single modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J. molec. Biol.86 (1974) 245–260.
Yarus, M., Translational efficiency of transfer RNAs: uses of an extended anticodon. Science218 (1982) 646–652.
Zelwer, C., Risler, J. L., and Brunie, S., Crystal structure ofEscherichia coli methionyl-tRNA synthetase at 2.5 Å resolution. J. molec. Biol.155 (1982) 63–81.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Söll, D. The accuracy of aminoacylation — ensuring the fidelity of the genetic code. Experientia 46, 1089–1096 (1990). https://doi.org/10.1007/BF01936918
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF01936918