Abstract
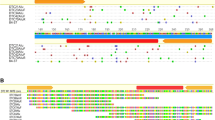
We report the cloning ofhermit, a member of thehAT family of transposable elements from the genome of the Australian sheep blowfly,Lucilia cuprina. Hermit is 2716 bp long and is 49% homologous to the autonomoushobo element,HFL1, at the nucleic acid level.Hermit has 15 bp terminal inverted repeats that share 10 bp with the terminal inverted repeats ofHFL1. Conceptual translation reveals a 583 residue open reading frame (ORF) that is 64% similar and 42% identical to theHFL1 ORF. However, the sequence of thehermit element contains two frameshifts within the putative ORF, indication thathermit is an inactive element. Analysis ofL. cuprina strains from within and outside Australia suggested thathermit is present as a single copy in all the genomes analysed.
Similar content being viewed by others
References
Atkinson, P. W., W. D. Warren & D. A. O'Brochta, 1993. The hobo transposable element ofDrosophila can be cross-mobilised in houseflies and excises like the Ac element of maize. Proc. Natl. Acad. Sci. USA 90: 9693–9697.
Blackman, R. K., M. M. Koehler, R. Grimaila & W. M. Gelbart, 1989. Identification of a fully-functional hobo transposable element and its use for germ-like transformation ofDrosophila. The EMBO Journal 8: 211–217.
Board, P., R. Russell, R. Marano & J. G. Oakeshott, 1994. Purification, molecular cloning and heterologous expression of a glutathione S-transferase from the Australian sheep blowfly (Lucilia cuprina). Biochem. Journal 299: 425–430.
Boussy, I. A. & G. Periquet, 1993. The transposable element hobo inDrosophila melanogaster and related species, pp 192–200 in Transposable Elements and Evolution, edited by J.F. McDonald. Kluwer Academic Publishers, Netherlands.
Calvi, B. R., T. J. Hong, S. D. Findley & W. M. Gelbart, 1991. Evidence for a common evolutionary origin of inverted repeat transposons inDrosophila and plants: hobo, Activator, and Tam3. Cell 66: 465–471.
Crozier, Y. C., S. Koulianos & R. H. Crozier, 1991. An improved test for Africanized honeybee mitochondrial DNA. Experientia 47: 968–969.
Daniels, S. B., A. Chovnick & I. A. Boussy, 1990. Distribution of hobo transposable elements in the genusDrosophila. Mol. Biol. Evol. 7: 589–606.
Fedoroff, N. V., 1989. Maize Transposable Elements, pp. 375–412 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology, Washington, D.C.
Elizur, A., A. T. Vacek & A. J. Howells, 1990. Cloning and characterisation of the white and topaz eye colour genes from the sheep blowflyLucilia cuprina. J. Mol. Evol. 30: 347–358.
Feldmar, S. & R. Kunze, 1991. The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. The EMBO Journal 10: 4003–4010.
Haring, M. A., C. M. T. Rommens, H. J. J. Nijkamp & J. Hille, 1991. The use of transgenic plants to understand transposition mechanisms and to develop transposon tagging strategies. Plant Molecular Biology 16: 449–461.
Hehl, R., W. K. F. Nacken, A. Krause, H. Saedler & H. Sommer, 1991. Structural analysis of Tam3, a transposable element fromAntirrhinum majus, reveals homologies to the Ac element from maize. Plant Molecular Biology 16: 369–371.
Jones, D. A., C. M. Thomas, K. E. Hammond-Kosack, P. J. Balint-Kurti & J. D. G. Jones, 1994. Isolation of the tomato Cf-9 Gene for resistance toCladosporium fulvum by transposon tagging. Science 266: 789–793.
Kaplan, N. L., R. R. Hudson & C. H. Langley, 1989. The ‘hitchhiking effect’ revisited. Genetics 123: 887–899.
Lidholm, D-A., A. R. Lohe & D. L. Hartl, 1993. The transposable element mariner mediates germline transformation inDrosophila melanogaster. Genetics 134: 859–868.
Maynard Smith, J. & J. Haigh, 1974. The hitchhiking effect of a favourable gene. Genet. Res. 23: 23–35.
Nasmyth, K. A., 1982. The regulation of yeast mating type. Chromatin structure by SIR: An action at a distance effecting both transcription and transposition. Cell 30: 567–578.
O'Brochta, D. A. & A. M. Handler, 1993. Prospects and Possibilities for Gene Transfer Techniques in Insects. pp. 451–488 in Molecular Approaches to Fundamental and Applied Entomology, edited by J. Oakeshott and M. J. Whitten. Springer-Verlag, New York Inc., N.Y.
O'Brochta, D. A., W. D. Warren, K. J. Saville & P. W. Atkinson, 1994. Interplasmid transposition ofDrosophila hobo elements in non-drosophilid insects. Mol. Gen. Genet. 244: 9–14.
Pascual, L. & G. Periquet, 1991. Distribution of hobo transposable elements in natural populations ofDrosophila melanogaster. Mol. Biol. Evol. 8: 282–296.
Perkins, H. D. & A. J. Howells, 1992. Genomic sequences with homology to the P element ofDrosophila melanogaster occur in the blowflyLucilia cuprina. Proc. Natl. Acad. Sci. USA 89: 10753–10757.
Pridmore, R. D., 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56: 309–312.
Rubin, G. M. & A. C. Spradling, 1982. Genetic transformation ofDrosophila with transposable element vectors. Science 218: 348–353.
Sambrook, J., E. F. Fritsch & T. Maniatis, 1989. Molecular Cloning: A laboratory manual; Cold Spring Harbour Laboratory Press, 3 Vols., Cold Spring Harbour, N.Y.
Sanger, F., S. Nicklen & A. R. Coulson, 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74: 5463–5467.
Simmons, G. M., 1992. Horizontal transfer of hobo transposable elements within theDrosophila melanogaster species complex: Evidence from DNA sequencing. Mol. Biol. Evol. 9: 1050–1060.
Streck, R. D., J. E. MacGaffey & S. K. Beckendorf, 1986. The structure of hobo transposable elements and their insertion sites. The EMBO Journal 5: 3615–3623.
Toung, Y. P. S., T. S. Hsieh & C. P. D. Tu, 1990.Drosophila glutathione S-transferase 1–1 shares a region of sequence homology with the maize glutathione S-transferase III. Proc. Natl. Acad. Sci. USA 87: 31–35.
Wang, J. Y., S. McCommas & M. Syvanen, 1991. Molecular cloning of a glutathione S-transferase overproduced in an insecticide resistant strain of the housefly (Musca domestica). Mol. Gen. Genet. 227: 260–266.
Warren, W. D., P. W. Atkinson & D. A. O'Brochta, 1994. The Hermes transposable element from the houseflyMusca domestica is a short inverted repeat-type element of the hobo, Ac and Tam3 (hAT) element family. Genet. Res. Camb. 64: 87–97.
Warren, W. D., P. W. Atkinson & D. A. O'Brochta, 1995. The Australian Bushfly,Musca vetustissima contains a sequence related to the transposons of the hobo, Ac and Tam3 family. Gene 154: 137–138.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Coates, C.J., Johnson, K.N., Perkins, H.D. et al. Thehermit transposable element of the Australian sheep blowfly,Lucilia cuprina, belongs to thehAT family of transposable elements. Genetica 97, 23–31 (1996). https://doi.org/10.1007/BF00132577
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00132577