Abstract
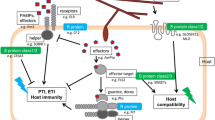
The bacterial speck disease of tomato has been developed as a model system to elucidate the molecular basis of specificity in plant-bacterial interactions and to study signal transduction events involved in the expression of plant disease resistance. We have employed a mutagenic approach to define the steps involved in the expression of disease resistance to the bacterial pathogen, Pseudomonas syringae pv. tomato (Pst) Eleven disease susceptible mutants have been identified and characterized twith altered recognition of Pst strains that express the avirulence gene avrPto. Genetic analysis of these mutants has revealed that they fall into two complementation groups. Five of the mutants map at the Pto locus, while six map a new locus that we have termed Prf. Further characterization of these mutants has revealed that the mutants that map at Pto are still sensitive to the insecticide fenthion, while the prf mutants are altered in resistance and also are insensitive to fenthion. Genetic mapping has also determined that the Prf locus maps near Pto. We are currently employing a map-based cloning strategy to isolate the Prf locus.
Similar content being viewed by others
References
Bennetzen, J. & J. Jones, 1992. Approaches and progress in molecular cloning of plant disease resistance genes. In: Genetic Engineering: Principles and Methods (in press).
Carland, F. & B. Staskawicz, 1993. Genetic characterization of the Pto locus of tomato: semi-dominance and segregation of resistance to Pseudomonas syringae pathovar tomato and sensitivity to the insecticide Fenthion. Mol. Gen. Genet. 239: 17–27.
De Wit, P.J., A.E. Hofman, G. Velthuis & J. Kuc, 1985. Isolation and characterization of an elicitor of necrosis isolated from intercellular fluids of compatible interactions of Cladosporium fulvum and tomato. Plant Phys. 77: 642–647.
Dixon, R.A. & C.J. Lamb, 1990. Molecular communication in interactions between plants and microbial pathogens. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 339–367.
Flor, H.H., 1947. Host-parasite interactions in flax rust-its genetics and other implications. Phytopathology 45: 680–685.
Gabriel, D. & B. Rolfe, 1990. Working models of specific recognition in plant-microbe interactions. Ann. Rev. Phytopathol. 28: 365–391.
Grimm, C. & N. Panopoulos, 1989. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several prokaryotic regulatory proteins. J. Bacteriol. 171: 5031–5038.
Innes, R.W., A.F. Bent, B.N. Knukel, S.R. Bisgrove & B.J. Staskawicz, 1993. Molecular characterization of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol. 175 (15): 4859–4869.
Johal, G.S. & S.P. Briggs, 1992. Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258: 985–987.
Keen, N.T., 1990. Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24: 447–463.
Keen, N.T., J.J. Sims, S. Midland, M. Yoder, F. Jurnak, H. Shen, C. Boyd, I. Yucel, J. Lorang & J. Murillo, 1993. Determinants of specificity in the interaction of plants with bacterial pathogens. In: Nester, EW & Verma, DPS (Ed.), Advances in Molecular Genetics of Plant-Microbe Interactions, pp. 211–220. Kluwer Academic Publishers, Dordrecht, The Netherlands.
Keen, N.T., S. Tamaki, D. Kobayashi, D. Gerhold, M. Stayton, H. Shen, S. Gold, J. Lorang, H. Thordal-Christensen, D. Dahlbeck & B. Staskawicz, 1990. Bacteria expressing avirulence gene D produce a specific elicitor of the soybean hypersensitive reaction. Mol. Plant-Microbe Interact. 3: 112–121.
Klement, Z., 1982. Hypersensitivity. In: Mount, M.S. & Lacy, G.H. (Ed.), Phytopathogenic Procaryotes, pp. 149–177. Academic Press, New York.
Laterrot, H. & J. Philouze, 1985. Susceptibility of the Pto plants to lebaycid insecticide: a tool for plant breeders? Tomato Genet. Coop. Rep. 35: 6.
Lindgren, P.B., R.C. Peet & N.J. Panopoulos, 1986. Gene cluster of Pseudomonas syringae pv. ‘phaseolicola’ controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J. Bacteriol. 168: 512–522.
Long, S. & B. Staskawicz, 1993. Prokaryotic plant parasites. Cell 73: 921–935.
Martin, G.B., S.H. Brommonschenkel, J. Chunwongse, A. Frary, M.W. Ganal, R. Spivey, T. Wu, E.D. Earle & S.D. Tanksley, 1993a. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436.
Martin, G.B., M.C.De Vicente & S.D. Tanksley, 1993b. Highresolution linkage analysis and physical characterization of the Pto bacterial resistance locus in tomato. Mol. Plant-Microbe Interact. 6: 26–34.
Martin, G.B., M.W. Ganal & S.D. Tanksley, 1992. Construction of a yeast artificial chromosome library of tomato and identification of cloned segments linked to two disease resistance loci. Mol. Gen. Genet. 233: 25–32.
Meeley, R.B., G.S. Johal, S.P. Briggs & J.D. Walton, 1992. A biochemical phenotype for a disease resistance gene of maize. Plant Cell 4: 71–77.
Pitblado, R.E. & B.H. MacNeill, 1983. Genetic basis of resistance to Pseudomonas syringae pv. tomato in field tomatoes. Can. J. Plant Pathol. 5: 251–255.
Reardon, J.T., C.A. Liljestrand-Golden, R.L. Dusenbery & P.D. Smith, 1987. Molecular analysis of diepoxybutane-induced mutations at the rosy locus of Drosophila melanogaster. Genetics 115: 323–331.
Ronald, P.C., J.M. Salmeron, F.C. Carland & B.J. Staskawicz, 1992. Cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 174: 1604–1611.
Salmeron, J.M., S.J. Barker, F.M. Carland, A.Y. Mehta & B.J. Staskawicz, 1994. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell 6: 511–520.
Salmeron, J.M. & B.J. Staskawicz, 1993. Molecular characterization and hrp-dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol. Gen. Genet. 239: 6–16.
Shirley, B.W., S. Hanley & H.M. Goodman, 1992. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347.
Sun, T.P., H.M. Goodman & F.M. Ausubel, 1992. Cloning of the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128.
Van Kan, J.A.L., G.F.J.M.Van den Ackerveken & P.J.G.M.De Wit, 1991. Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant-Microbe Interact. 4: 52–59.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Salmeron, J., Rommens, C., Barker, S. et al. Genetic dissection of bacterial speck disease resistance in tomato. Euphytica 79, 195–200 (1994). https://doi.org/10.1007/BF00022519
Issue Date:
DOI: https://doi.org/10.1007/BF00022519